Investigating the time to blood culture positivity: why does it take so long?
- PMID: 39757997
- PMCID: PMC11701752
- DOI: 10.1099/jmm.0.001942
Investigating the time to blood culture positivity: why does it take so long?
Abstract
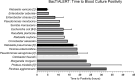
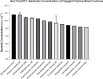
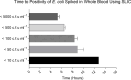
Introduction. Bloodstream infections (BSIs) are one of the most serious infections investigated by microbiologists. However, the time to detect a BSI fails to meet the rapidity required to inform clinical decisions in real time.Gap Statement. Blood culture (BC) is considered the gold standard for diagnosing bloodstream infections. However, the time to blood culture positivity can be lengthy. Underpinning this is the reliance on bacteria replicating to a high concentration, which is necessary for the detection using routine blood culture systems. To improve the diagnosis and management of patients with BSIs, more sensitive detection methods are required.Aim. The study aimed to answer key questions addressing the delay in BSI detection and whether the time to BSI detection could be expedited using a Scattered Light Integrated Collection (SLIC) device.Methodology. A proof-of-concept study was conducted to compare the time to positivity (TTP) of Gram-negative BCs flagging positive on BacT/ALERT with an SLIC device. An SLIC device was utilized to compare the TTP of the most prevalent BSI pathogens derived from nutrient broth and BC, the influence of bacterial load on TTP and the TTP directly from whole blood. Additionally, the overall turnaround time (TAT) of SLIC was compared with that of a standard hospital workflow.Results. Most pathogens tested took significantly longer to replicate when derived from BC than from nutrient medium. The median TTP of Gram-negative BC on BacT/ALERT was 13.56 h with a median bacterial load of 6.4×109 c.f.u. ml-1. All pathogens (7/7) derived from BC at a concentration of 105 c.f.u. ml-1 were detectable in under 70 min on SLIC. Decreasing Escherichia coli BC concentration from 105 to 102 c.f.u. ml-1 increased the TTP of SLIC from 15 to 85 min. Direct BSI detection from whole blood on SLIC demonstrated a 76% reduction in TAT when compared with the standard hospital workflow.Conclusion. An SLIC device significantly reduced the TTP of common BSI pathogens. The application of this technology could have a major impact on the detection and management of BSI.
Keywords: blood culture; bloodstream infection; rapid diagnostics.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures



Similar articles
-
Rapid determination of antimicrobial susceptibility of Gram-negative bacteria from clinical blood cultures using a scattered light-integrated collection device.J Med Microbiol. 2024 Feb;73(2). doi: 10.1099/jmm.0.001812. J Med Microbiol. 2024. PMID: 38415707
-
Clinical diagnostic performance of droplet digital PCR for pathogen detection in patients with Escherichia coli bloodstream infection: a prospective observational study.BMC Infect Dis. 2025 Jan 6;25(1):22. doi: 10.1186/s12879-024-10396-y. BMC Infect Dis. 2025. PMID: 39757158 Free PMC article.
-
Comparative evaluation of BacT/ALERT VIRTUO and BACTEC FX400 blood culture systems for the detection of bloodstream infections.Microbiol Spectr. 2025 Jan 7;13(1):e0185024. doi: 10.1128/spectrum.01850-24. Epub 2024 Nov 29. Microbiol Spectr. 2025. PMID: 39611835 Free PMC article.
-
The correct blood volume for paediatric blood cultures: a conundrum?Clin Microbiol Infect. 2020 Feb;26(2):168-173. doi: 10.1016/j.cmi.2019.10.006. Epub 2019 Oct 23. Clin Microbiol Infect. 2020. PMID: 31654793 Review.
-
Bloodstream infections - Standard and progress in pathogen diagnostics.Clin Microbiol Infect. 2020 Feb;26(2):142-150. doi: 10.1016/j.cmi.2019.11.017. Epub 2019 Nov 22. Clin Microbiol Infect. 2020. PMID: 31760113 Review.
Cited by
-
CGRP, PD-1 and PD-L1 as Biomarkers for PICC-Related Bloodstream Infections in Breast Cancer Patients.Technol Cancer Res Treat. 2025 Jan-Dec;24:15330338251342877. doi: 10.1177/15330338251342877. Epub 2025 May 21. Technol Cancer Res Treat. 2025. PMID: 40398661 Free PMC article.
-
The Clinical Utility and Stewardship Challenges of Time to Blood Culture Positivity.Hosp Pharm. 2025 Aug 27:00185787251364488. doi: 10.1177/00185787251364488. Online ahead of print. Hosp Pharm. 2025. PMID: 40894300 Free PMC article. No abstract available.
-
Advancing rapid diagnostics for bloodstream infections: a perspective on scattered light integrated collection technology.J Med Microbiol. 2025 Mar;74(3):001973. doi: 10.1099/jmm.0.001973. J Med Microbiol. 2025. PMID: 40072045 Free PMC article. No abstract available.
References
-
- Surrey and Sussex Healthcare NHS Trust Microbiology laboratory user’s manual. 2017. [9-January-2018]. https://www.surreyandsussex.nhs.uk/wp- content/uploads/2013/05/Microbiol... accessed.
-
- Alder Hey Children’s NHS Foundation Trust A user’s guide to the medical microbiology department [accessed January 2018] 2017 http://www.alderhey.nhs.uk/wp- content/uploads/Microbiology-Lab-Handbook...
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

