Identification and comparison of N-glycome profiles from common dietary protein sources
- PMID: 39758068
- PMCID: PMC11699095
- DOI: 10.1016/j.fochx.2024.102025
Identification and comparison of N-glycome profiles from common dietary protein sources
Abstract
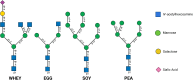
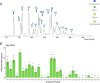
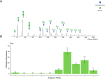
The N-glycomes of bovine whey, egg white, pea, and soy protein isolates are described here. N-glycans from four protein isolates were analyzed by HILIC high performance liquid chromatography and quadrupole time-of-flight tandem mass spectrometry (HILIC-FLD-QTOF-MS/MS). In total, 33 N-glycans from bovine whey and egg white and 10 N-glycans from soy and pea glycoproteins were identified. The type of N-glycans per glycoprotein source were attributable to differences in biosynthetic glycosylation pathways. Animal glycoprotein sources favored a combination of complex and hybrid glycan configurations, while the plant proteins were dominated by oligomannosidic N-glycans. Bovine whey glycoprotein isolate contained the most diverse N-glycans by monosaccharide composition as well as structure, while plant sources such as pea and soy glycoprotein isolates contained an overlap of oligomannosidic N-glycans. The results suggest N-glycan structure and composition is dependent on the host organism which are driven by the differences in N-glycan biosynthetic pathways.
Keywords: Glycan; Mass spectrometry; Microbiome; N-glycan; N-glycome; Protein.
© 2024 The Authors.
Conflict of interest statement
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Steven Frese reports financial support was provided by the United States Department of Agriculture's National Institute of Food and Agriculture. The other authors have no known competing financial interests have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



References
-
- Anderson M.J. A new method for non-parametric multivariate analysis of variance. Austral Ecology. 2001;26(1):32–46. doi: 10.1111/j.1442-9993.2001.01070.pp.x. - DOI
-
- Barboza M., Pinzon J., Wickramasinghe S., Froehlich J.W., Moeller I., Smilowitz J.T.…Lebrilla C.B. Glycosylation of human milk lactoferrin exhibits dynamic changes during early lactation enhancing its role in pathogenic bacteria-host interactions *. Molecular & Cellular Proteomics. 2012;11(6) doi: 10.1074/mcp.M111.015248. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

