Diagnostic Impact of FISHseq as a New Pathologic Criterion for Endocarditis According to the Duke Criteria
- PMID: 39758740
- PMCID: PMC11697105
- DOI: 10.1093/ofid/ofae716
Diagnostic Impact of FISHseq as a New Pathologic Criterion for Endocarditis According to the Duke Criteria
Abstract
Background: For clinicians treating patients with infective endocarditis (IE), identifying the causative microorganisms poses a critical diagnostic challenge. Standard techniques including blood and heart valve cultures often yield inconclusive results. According to the recent 2023 Duke-ISCVID Criteria, molecular methods represent potent tools to enhance this aspect of IE diagnostics and guide subsequent therapeutic strategies.
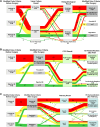
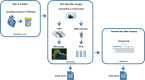
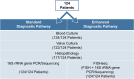
Methods: We retrospectively analyzed data from 124 consecutive patients who underwent heart valve surgery due to suspected IE at München Klinik Bogenhausen. The standard diagnostic pathway, which included blood culture, valve culture, histopathological analysis, and polymerase chain reaction (PCR)/sequencing, was compared with the enhanced diagnostic pathway, which included fluorescence in situ hybridization + PCR/sequencing (FISHseq) instead of PCR/sequencing alone. The aim of this study was to assess the added value of combining standard diagnostics with molecular methods such as PCR/sequencing or FISHseq for the diagnosis of IE and the potential impact on therapy.
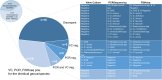
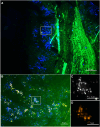
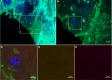
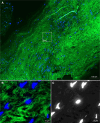
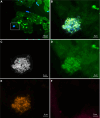
Results: Standard diagnostic methods and PCR/sequencing yielded inconclusive results in 57/124 cases (46.0%). FISHseq provided an added value for diagnostics in 79/124 cases (63.7%) and potentially would have impacted therapy in 95/124 (76.6%) of cases. By adding data through direct visualization and characterization of microorganisms, FISHseq reduced the number of inconclusive cases by 86.0%.
Conclusions: The comparison of 2 molecular diagnostic tools for IE from the same heart valve emphasizes the value of molecular methods including molecular imaging by FISH for IE diagnostics and supports the 2023 Duke-ISCVID Criteria.
Keywords: 2023 Duke-ISCVID Criteria; FISHseq; fluorescence in situ hybridization; infective endocarditis; molecular imaging.
© The Author(s) 2024. Published by Oxford University Press on behalf of Infectious Diseases Society of America.
Conflict of interest statement
Potential conflicts of interest. J.K. is CEO of MoKi Analytics GmbH. A.M. and J.K. are shareholders of MoKi Analytics GmbH. A.M. is the owner of the private practice Moter Diagnostics. All other authors report no potential conflicts.
Figures


References
-
- Cahill TJ, Baddour LM, Habib G, et al. Challenges in infective endocarditis. J Am Coll Cardiol 2017; 69:325–44. - PubMed
-
- Coffey S, Roberts-Thomson R, Brown A, et al. Global epidemiology of valvular heart disease. Nat Rev Cardiol 2021; 18:853–64 - PubMed
-
- Li JS, Sexton DJ, Mick N, et al. Proposed modifications to the Duke Criteria for the diagnosis of infective endocarditis. Clin Infect Dis 2000; 30:633–8. - PubMed
LinkOut - more resources
Full Text Sources

