Integration of pharmacogenetic data in epic genomic module drives clinical decision support alerts
- PMID: 39759455
- PMCID: PMC11695119
- DOI: 10.3389/fphar.2024.1458095
Integration of pharmacogenetic data in epic genomic module drives clinical decision support alerts
Abstract
Introduction: The Precision Medicine Program (PMP) at the University of Florida (UF) focuses on advancing pharmacogenomics (PGx) to improve patient care.
Methods: The UF PMP, in collaboration with the UF Health Pathology Laboratory (UFHPL), utilized Health Level Seven (HL7) standards to integrate PGx data into Epic's Genomic Module to enhance the management and utilization of PGx data in clinical practice.
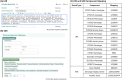
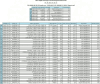
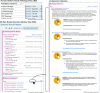
Results: A key feature of the Genomic Module is the introduction of genomic indicators-innovative tools that flag actionable genetic information directly within the electronic health record (EHR). These indicators enable the effective presentation of phenotypic information and, when leveraged with existing clinical decision support (CDS) alerts, help provide timely and informed therapeutic decisions based on genomic data.
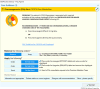
Discussion: This advancement represents a significant shift in the utilization of genetic data, moving beyond traditional PDF reports to provide a comprehensive understanding of PGx data. Ultimately, this integration empowers healthcare providers with genomics-guided recommendations, enhancing precision and personalization in patient care, contributing significantly to the advancement of personalized medicine.
Keywords: clinical decision support systems; electronic health records (EHR); genomics; health informatics; health level seven (HL7); pharmacogenetics; precision medicine.
Copyright © 2024 Newsom, Hall, Martinez, Nelson, Starostik and Nguyen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Caudle K. E., Dunnenberger H. M., Freimuth R. R., Peterson J. F., Burlison J. D., Whirl-Carrillo M., et al. (2017). Standardizing terms for clinical pharmacogenetic test results: consensus terms from the Clinical Pharmacogenetics Implementation Consortium (CPIC). Genet. Med. 19, 215–223. 10.1038/gim.2016.87 - DOI - PMC - PubMed
-
- Chamala S., Majety S., Mishra S. N., Newsom K. J., Gothi S. R., Walton N. A., et al. (2020). Indispensability of clinical bioinformatics for effective implementation of genomic medicine in Pathology laboratories. ACI open 04, e167–e172. 10.1055/s-0040-1721480 - DOI
LinkOut - more resources
Full Text Sources

