Unraveling metabolic signatures in SARS-CoV-2 variant infections using multiomics analysis
- PMID: 39759510
- PMCID: PMC11697598
- DOI: 10.3389/fimmu.2024.1473895
Unraveling metabolic signatures in SARS-CoV-2 variant infections using multiomics analysis
Abstract
Introduction: The emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants, notably delta and omicron, has significantly accelerated the global pandemic, worsening conditions worldwide. However, there is a lack of research concerning the molecular mechanisms related to immune responses and metabolism induced by these variants.
Methods: Here, metabolomics combined with transcriptomics was performed to elucidate the immunometabolic changes in the lung of hamsters infected with delta and omicron variants.
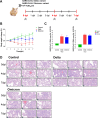
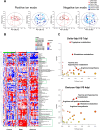
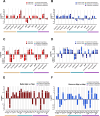
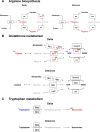
Results: Both variants caused acute inflammation and lung pathology in intranasally infected hamsters. Principal component analysis uncovered the delta variant significantly altered lung metabolite levels between the pre- and post-infection states. Additionally, metabolic pathways determined by assessment of metabolites and genes in lung revealed significant alterations in arginine biosynthesis, glutathione metabolism, and tryptophan metabolism upon infection with both variants and closely linked to inflammatory cytokines, indicating immune activation and oxidative stress in response to both variants. These metabolic changes were also evident in the serum, validating the presence of systemic alterations corresponding to those identified in lung. Notably, the delta variant induced a more robust metabolic regulation than the omicron variant.
Discussion: The study suggests that multi-omics is a valuable approach for understanding immunometabolic responses to infectious diseases, and providing insights for effective treatment strategies.
Keywords: SARS-CoV-2; immune response; metabolic pathway; metabolomics; transcriptomics.
Copyright © 2024 Lee, Lee, Lyoo, Shin, Shin, Kim, Yang, Kim, Lee and Hwang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

