uORF-targeting steric block antisense oligonucleotides do not reproducibly increase RNASEH1 expression
- PMID: 39759875
- PMCID: PMC11697566
- DOI: 10.1016/j.omtn.2024.102406
uORF-targeting steric block antisense oligonucleotides do not reproducibly increase RNASEH1 expression
Abstract
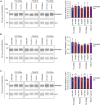
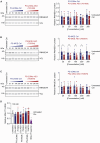
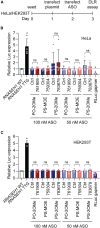
Upstream open reading frames (uORFs) are cis-regulatory motifs that are predicted to occur in the 5' UTRs of the majority of human protein-coding transcripts and are typically associated with translational repression of the downstream primary open reading frame (pORF). Interference with uORF activity provides a potential mechanism for targeted upregulation of the expression of specific transcripts. It was previously reported that steric block antisense oligonucleotides (ASOs) can bind to and mask uORF start codons to inhibit translation initiation, and thereby disrupt uORF-mediated gene regulation. Given the relative maturity of the oligonucleotide field, such a uORF blocking mechanism might have widespread therapeutic utility. Here, we re-synthesized three of the most potent ASOs targeting the RNASEH1 uORF described in a study by Liang et al. and investigated their potential for RNASEH1 protein upregulation, with care taken to replicate the conditions of the original study. No upregulation (of endogenous or reporter protein expression) was observed with any of the oligonucleotides tested at doses ranging from 25 to 300 nM. Conversely, we observed downregulation of expression in some instances. We conclude that previously described RNASEH1 uORF-targeting steric block ASOs are incapable of upregulating pORF protein expression in our hands.
Keywords: MT: Oligonucleotides: Therapies and Applications; RNASEH1; antisense oligonucleotides; steric block ASO; uORF; upstream open reading frame.
© 2024 The Authors.
Conflict of interest statement
T.C.R., M.J.A.W., and B.H. have filed a patent related to a uORF-targeting ASO technology. T.C.R., M.J.A.W., N.S., and B.H. are founders and shareholders in Orfonyx Bio Ltd., a biotechnology spin-out company that aims to utilize uORF-targeting technologies for therapeutics development. N.S. is an employee of Orfonyx Bio. T.C.R. and M.J.A.W. are consultants for Orfonyx Bio.
Figures
Similar articles
-
Secondary structures that regulate mRNA translation provide insights for ASO-mediated modulation of cardiac hypertrophy.bioRxiv [Preprint]. 2023 Jun 15:2023.06.15.545153. doi: 10.1101/2023.06.15.545153. bioRxiv. 2023. Update in: Nat Commun. 2023 Oct 3;14(1):6166. doi: 10.1038/s41467-023-41799-1. PMID: 37397986 Free PMC article. Updated. Preprint.
-
Specific Increase of Protein Levels by Enhancing Translation Using Antisense Oligonucleotides Targeting Upstream Open Frames.Adv Exp Med Biol. 2017;983:129-146. doi: 10.1007/978-981-10-4310-9_9. Adv Exp Med Biol. 2017. PMID: 28639196 Review.
-
Secondary structures that regulate mRNA translation provide insights for ASO-mediated modulation of cardiac hypertrophy.Nat Commun. 2023 Oct 3;14(1):6166. doi: 10.1038/s41467-023-41799-1. Nat Commun. 2023. PMID: 37789015 Free PMC article.
-
Translation efficiency of mRNAs is increased by antisense oligonucleotides targeting upstream open reading frames.Nat Biotechnol. 2016 Aug;34(8):875-80. doi: 10.1038/nbt.3589. Epub 2016 Jul 11. Nat Biotechnol. 2016. PMID: 27398791
-
Translational Regulation by Upstream Open Reading Frames and Human Diseases.Adv Exp Med Biol. 2019;1157:99-116. doi: 10.1007/978-3-030-19966-1_5. Adv Exp Med Biol. 2019. PMID: 31342439 Review.
References
LinkOut - more resources
Full Text Sources

