A sort and sequence approach to dissect heterogeneity of response to a self-amplifying RNA vector in a novel human muscle cell line
- PMID: 39759876
- PMCID: PMC11700297
- DOI: 10.1016/j.omtn.2024.102400
A sort and sequence approach to dissect heterogeneity of response to a self-amplifying RNA vector in a novel human muscle cell line
Abstract
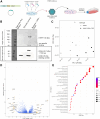
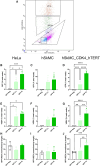
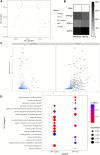
Self-amplifying RNA (saRNA) is an extremely promising platform because it can produce more protein for less RNA. We used a sort and sequence approach to identify host cell factors associated with transgene expression from saRNA; the hypothesis was that cells with different expression levels would have different transcriptomes. We tested this in CDK4/hTERT immortalized human muscle cells transfected with Venezuelan equine encephalitis virus (VEEV)-derived saRNA encoding GFP. Cells with the highest expression levels had very high levels of transgene mRNA (5%-10% total reads); the cells sorted with low and negative levels of GFP protein also had detectable levels of both VEEV and GFP RNA. To understand host responses, we performed RNA sequencing. Differentially expressed gene (DEG) patterns varied with GFP expression; GFP high cells had many more DEGs, which were associated with protein synthesis and cell metabolism. Comparing profiles by an unsupervised approach revealed that negative cells expressed higher levels of cell-intrinsic immunity genes such as IFIT1, MX1, TLR3, and MyD88. To explore the role of interferon, cells were treated with the Jak inhibitor ruxolitinib. This reduced the number of DEGs, but differences between cells sorted by expression level remained. These studies demonstrate the complex interplay of factors, some immune related, affecting saRNA transgenes.
Keywords: MT: Delivery Strategies; RNA; alphavirus; expression; innate immunity; self-amplifying; vaccine.
© 2024 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures






References
-
- Pepini T., Pulichino A.M., Carsillo T., Carlson A.L., Sari-Sarraf F., Ramsauer K., Debasitis J.C., Maruggi G., Otten G.R., Geall A.J., et al. Induction of an IFN-Mediated Antiviral Response by a Self-Amplifying RNA Vaccine: Implications for Vaccine Design. J. Immunol. 2017;198:4012–4024. - PMC - PubMed
-
- Guy C., Bowie A.G. Recent insights into innate immune nucleic acid sensing during viral infection. Curr. Opin. Immunol. 2022;78 - PubMed
-
- Shirota H., Petrenko L., Hong C., Klinman D.M. Potential of transfected muscle cells to contribute to DNA vaccine immunogenicity. J. Immunol. 2007;179:329–336. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

