SARS-CoV-2 Productively Infects Human Hepatocytes and Induces Cell Death
- PMID: 39760326
- PMCID: PMC11702151
- DOI: 10.1002/jmv.70156
SARS-CoV-2 Productively Infects Human Hepatocytes and Induces Cell Death
Abstract
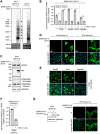
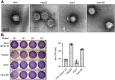
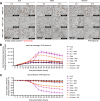
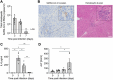
SARS-CoV-2 infection is accompanied by elevated liver enzymes, and patients with pre-existing liver conditions experience more severe disease. While it was known that SARS-CoV-2 infects human hepatocytes, our study determines the mechanism of infection, demonstrates viral replication and spread, and highlights direct hepatocyte damage. Viral replication was readily detectable upon infection of primary human hepatocytes and hepatoma cells with the ancestral SARS-CoV-2, Delta, and Omicron variants. Hepatocytes express the SARS-CoV-2 receptor ACE2 and the host cell protease TMPRSS2, and knocking down ACE2 and TMPRSS2 impaired SARS-CoV-2 infection. Progeny viruses released from infected hepatocytes showed the typical coronavirus morphology by electron microscopy and proved infectious when transferred to fresh cells, indicating that hepatocytes can contribute to virus spread. Importantly, SARS-CoV-2 infection rapidly induced hepatocyte death in a replication-dependent fashion, with the Omicron variant showing faster onset but less extensive cell death. C57BL/6 wild-type mice infected with a mouse-adapted SARS-CoV-2 strain showed high levels of viral RNA in liver and lung tissues. ALT peaked when viral RNA was cleared from the liver. Liver histology revealed profound tissue damage and immune cell infiltration, indicating that direct cytopathic effects of SARS-CoV-2 and immune-mediated killing of infected hepatocytes contribute to liver pathology.
Keywords: ACE2; COVID‐19; SARS‐CoV‐2; TMPRSS2; hepatocytes; liver; tropism.
© 2025 The Author(s). Journal of Medical Virology published by Wiley Periodicals LLC.
Conflict of interest statement
UP received grants from SCG Cell Therapy, and VirBio and personal fees from Abbott, Abbvie, Arbutus, Gilead, GSK, Leukocare, J&J, Roche, MSD, Sanofi, Sobi and Vaccitech. UP is a cofounder and share‐holder of SCG Cell Therapy. The other authors declare no conflicts of interest. CAJ reports research funding from the German Center for Lung Research and an issued patent “Bradykinin B2 receptor antagonist for treatment of SARS‐CoV2‐infection” (EP3909646A1).
Figures

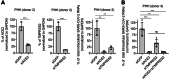
References
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

