Autonomous Nucleic Acid and Protein Nanocomputing Agents Engineered to Operate in Living Cells
- PMID: 39760461
- PMCID: PMC11757000
- DOI: 10.1021/acsnano.4c13663
Autonomous Nucleic Acid and Protein Nanocomputing Agents Engineered to Operate in Living Cells
Abstract
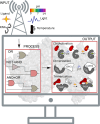
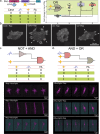
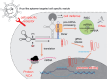
In recent years, the rapid development and employment of autonomous technology have been observed in many areas of human activity. Autonomous technology can readily adjust its function to environmental conditions and enable an efficient operation without human control. While applying the same concept to designing advanced biomolecular therapies would revolutionize nanomedicine, the design approaches to engineering biological nanocomputing agents for predefined operations within living cells remain a challenge. Autonomous nanocomputing agents made of nucleic acids and proteins are an appealing idea, and two decades of research has shown that the engineered agents act under real physical and biochemical constraints in a logical manner. Throughout all domains of life, nucleic acids and proteins perform a variety of vital functions, where the sequence-defined structures of these biopolymers either operate on their own or efficiently function together. This programmability and synergy inspire massive research efforts that utilize the versatility of nucleic and amino acids to encode functions and properties that otherwise do not exist in nature. This Perspective covers the key concepts used in the design and application of nanocomputing agents and discusses potential limitations and paths forward.
Keywords: directed evolution; nanocomputing agents; nucleic acid nanoparticles; proteins; rational design.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


References
-
- Ezenkwu C. P.; Starkey A. In Machine Autonomy: Definition, Approaches, Challenges and Research Gaps, Intelligent Computing; Arai K.; Bhatia R.; Kapoor S., Eds.; Springer International Publishing: Cham, 2019; pp 335–358.
-
- Abramson J. S.; Palomba M. L.; Gordon L. I.; Lunning M. A.; Wang M.; Arnason J.; Mehta A.; Purev E.; Maloney D. G.; Andreadis C.; Sehgal A.; Solomon S. R.; Ghosh N.; Albertson T. M.; Garcia J.; Kostic A.; Mallaney M.; Ogasawara K.; Newhall K.; Kim Y.; Li D.; Siddiqi T. Lisocabtagene maraleucel for patients with relapsed or refractory large B-cell lymphomas (TRANSCEND NHL 001): a multicentre seamless design study. Lancet 2020, 396 (10254), 839–852. 10.1016/S0140-6736(20)31366-0. - DOI - PubMed
-
- Munshi N. C.; Anderson L. D. Jr.; Shah N.; Madduri D.; Berdeja J.; Lonial S.; Raje N.; Lin Y.; Siegel D.; Oriol A.; Moreau P.; Yakoub-Agha I.; Delforge M.; Cavo M.; Einsele H.; Goldschmidt H.; Weisel K.; Rambaldi A.; Reece D.; Petrocca F.; Massaro M.; Connarn J. N.; Kaiser S.; Patel P.; Huang L.; Campbell T. B.; Hege K.; San-Miguel J. Idecabtagene Vicleucel in Relapsed and Refractory Multiple Myeloma. N Engl J. Med. 2021, 384 (8), 705–716. 10.1056/NEJMoa2024850. - DOI - PubMed
-
- Locatelli F.; Thompson A. A.; Kwiatkowski J. L.; Porter J. B.; Thrasher A. J.; Hongeng S.; Sauer M. G.; Thuret I.; Lal A.; Algeri M.; Schneiderman J.; Olson T. S.; Carpenter B.; Amrolia P. J.; Anurathapan U.; Schambach A.; Chabannon C.; Schmidt M.; Labik I.; Elliot H.; Guo R.; Asmal M.; Colvin R. A.; Walters M. C. Betibeglogene Autotemcel Gene Therapy for Non-beta(0)/beta(0) Genotype beta-Thalassemia. N Engl J. Med. 2022, 386 (5), 415–427. 10.1056/NEJMoa2113206. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

