Rapidly spreading Enterobacterales with OXA-48-like carbapenemases
- PMID: 39760498
- PMCID: PMC11837536
- DOI: 10.1128/jcm.01515-24
Rapidly spreading Enterobacterales with OXA-48-like carbapenemases
Abstract
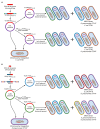
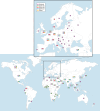
Enterobacterales (mostly Klebsiella pneumoniae, Escherichia coli) with OXA-48-like carbapenemases (e.g., OXA-48, -181, -232, -244) are undermining the global efficiency of carbapenem therapy. In the Middle East, North Africa, and some European countries, OXA-48-like carbapenemases are the most common types of carbapenemases among Enterobacterales. Currently, OXA-48 is endemic in the Middle East, North Africa, Spain, France, and Belgium; OXA-181 is endemic in Sub-Saharan Africa and the Indian Subcontinent, while OXA-232 has been increasing in the Indian Subcontinent. European countries (e.g., Germany, Denmark, Switzerland, France) are experiencing community outbreaks with E. coli ST38 that produce OXA-244, and these strains have been introduced into Norwegian, Polish, and Czech hospitals. The global ascendancy of OXA-48-like genes is due to the combination of carbapenemases with horizontal spread through promiscuous plasmids (e.g., IncL, IncX3, ColE2) and vertical spread with certain high-risk multidrug-resistant clones (e.g., K. pneumoniae ST14, ST15, ST147, ST307; E. coli ST38, ST410). This is a powerful "gene survival strategy" that has assisted with the survival of OXA-48-like genes in different environments including the community setting. The laboratory diagnosis is complex; therefore, bacteria with "difficult to detect" variants (e.g., OXA-244, OXA-484) are likely underreported and are spreading silently "beneath the radar" in hospital and community settings. K. pneumoniae and E. coli with OXA-48-like carbapenemases are forces to be reckoned with.
Keywords: OXA-48-like; carbapenemases; epidemiology; laboratory detection.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

