Haplotype-resolved genome of a papeda provides insights into the geographical origin and evolution of Citrus
- PMID: 39760510
- PMCID: PMC11814927
- DOI: 10.1111/jipb.13819
Haplotype-resolved genome of a papeda provides insights into the geographical origin and evolution of Citrus
Abstract
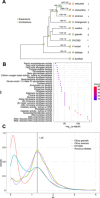
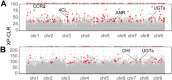
The publication of several high-quality genomes has contributed greatly to clarifying the evolution of citrus. However, due to their complex genetic backgrounds, the origins and evolution of many citrus species remain unclear. We assembled de novo the 294-Mbp chromosome-level genome of a more than 200-year-old primitive papeda (DYC002). Comparison between the two sets of homologous chromosomes of the haplotype-resolved genome revealed 1.2% intragenomic variations, including 1.75 million SNPs, 149,471 insertions and 154,215 deletions. Using this genome as a reference, we resequenced and performed population and phylogenetic analyses of 378 representative citrus accessions. Our study confirmed that the primary origin center of core Citrus species is in South China, particularly in the Himalaya-Hengduan Mountains. Papeda species are an ancient Citrus type compared with C. ichangensis. We found that the evolution of the Citrus genus followed two radiations through two routes (to East China and Southeast Asia) along river systems. Evidence for the origin and evolution of some individual citrus species was provided. Papeda probably played an important role in the origins of Australian finger lime, citrons, Honghe papeda and pummelos; Ichang papeda originated from Yuanjiang city of Yunnan Province, China, and C. mangshanensis has a close relationship with kumquat and Ichang papeda. Moreover, the Hunan and Guangdong Provinces of China are predicted to be the origin center of mandarin, sweet orange and sour orange. Additionally, our study revealed that fruit bitterness was significantly selected against during citrus domestication. Taken together, this study provides new insight into the origin and evolution of citrus species and may serve as a valuable genomic resource for citrus breeding and improvement.
Keywords: citrus; domestication; evolution; haplotype‐resolved genome; papeda.
© 2025 The Author(s). Journal of Integrative Plant Biology published by John Wiley & Sons Australia, Ltd on behalf of Institute of Botany, Chinese Academy of Sciences.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
Transcriptome and metabolome atlas reveals contributions of sphingosine and chlorogenic acid to cold tolerance in Citrus.Plant Physiol. 2024 Sep 2;196(1):634-650. doi: 10.1093/plphys/kiae327. Plant Physiol. 2024. PMID: 38875157
-
Phylogenetic origin of limes and lemons revealed by cytoplasmic and nuclear markers.Ann Bot. 2016 Apr;117(4):565-83. doi: 10.1093/aob/mcw005. Epub 2016 Mar 4. Ann Bot. 2016. PMID: 26944784 Free PMC article.
-
Characterization of the complete chloroplast genome of Yuanjiang wild Ichang papeda (Citrus cavaleriei) in China.Mitochondrial DNA B Resour. 2020 Sep 16;5(3):3349-3350. doi: 10.1080/23802359.2020.1820397. Mitochondrial DNA B Resour. 2020. PMID: 33458165 Free PMC article.
-
Etiology of three recent diseases of citrus in São Paulo State: sudden death, variegated chlorosis and huanglongbing.IUBMB Life. 2007 Apr-May;59(4-5):346-54. doi: 10.1080/15216540701299326. IUBMB Life. 2007. PMID: 17505974 Review.
-
Genomic insights into citrus domestication and its important agronomic traits.Plant Commun. 2020 Dec 30;2(1):100138. doi: 10.1016/j.xplc.2020.100138. eCollection 2021 Jan 11. Plant Commun. 2020. PMID: 33511347 Free PMC article. Review.
Cited by
-
Unlocking the Sublime: A Review of Native Australian Citrus Species.Foods. 2025 Jul 9;14(14):2425. doi: 10.3390/foods14142425. Foods. 2025. PMID: 40724244 Free PMC article. Review.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

