Single-cell RNA-sequencing and genome-wide Mendelian randomisation along with abundant machine learning methods identify a novel B cells signature in gastric cancer
- PMID: 39760915
- PMCID: PMC11703799
- DOI: 10.1007/s12672-025-01759-1
Single-cell RNA-sequencing and genome-wide Mendelian randomisation along with abundant machine learning methods identify a novel B cells signature in gastric cancer
Abstract
Background: Gastric cancer (GC) has a poor prognosis, considerable cellular heterogeneity, and ranks fifth among malignant tumours. Understanding the tumour microenvironment (TME) and intra-tumor heterogeneity (ITH) may lead to the development of novel GC treatments.
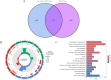
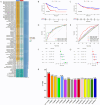
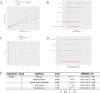
Methods: The single-cell RNA sequencing (scRNA-seq) dataset was obtained from the Gene Expression Omnibus (GEO) database, where diverse immune cells were isolated and re-annotated based on cell markers established in the original study to ascertain their individual characteristics. We conducted a weighted gene co-expression network analysis (WGCNA) to identify genes with a significant correlation to GC. Utilising bulk RNA sequencing data, we employed machine learning integration methods to train specific biomarkers for the development of novel diagnostic combinations. A two-sample Mendelian randomisation study was performed to investigate the causal effect of biomarkers on gastric cancer (GC). Ultimately, we utilised the DSigDB database to acquire associations between signature genes and pharmaceuticals.
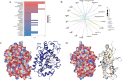
Results: The 18 genes that made up the signature were as follows: ZFAND2A, PBX4, RAMP2, NNMT, RNASE1, CD93, CDH5, NFKBIE, VWF, DAB2, FAAH2, VAT1, MRAS, TSPAN4, EPAS1, AFAP1L1, DNM3. Patients were categorised into high-risk and low-risk groups according to their risk scores. Individuals in the high-risk cohort exhibited a dismal outlook. The Mendelian randomisation study demonstrated that individuals with a genetic predisposition for elevated NFKBIE levels exhibited a heightened likelihood of acquiring GC. Molecular docking indicates that gemcitabine and chloropyramine may serve as effective therapeutics against NFKBIE.
Conclusions: We developed and validated a signature utilising scRNA-seq and bulk sequencing data from gastric cancer patients. NFKBIE may function as a novel biomarker and therapeutic target for GC.
Keywords: Bioinformatics; Diagnostic biomarker; Gastric cancer; Machine learning; Single-cell RNA sequencing.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Competing interests: The authors declare no competing interests.
Figures




References
-
- Smyth EC, et al. Gastric cancer. Lancet. 2020;396(10251):635–48. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
