Characterizing progenitor cells in developing and injured spinal cord: Insights from single-nucleus transcriptomics and lineage tracing
- PMID: 39761400
- PMCID: PMC11745359
- DOI: 10.1073/pnas.2413140122
Characterizing progenitor cells in developing and injured spinal cord: Insights from single-nucleus transcriptomics and lineage tracing
Abstract
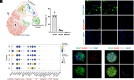
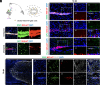
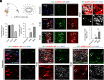
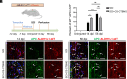
Various mature tissue-resident cells exhibit progenitor characteristics following injury. However, the existence of endogenous stem cells with multiple lineage potentials in the adult spinal cord remains a compelling area of research. In this study, we present a cross-species investigation that extends from development to injury. We used single-nucleus transcriptomic sequencing and genetic lineage tracing to characterize neural cells in the spinal cord. Our findings show that ciliated ependymal cells lose neural progenitor gene signatures and proliferation ability following the differentiation of NPCs within the ventricular zone. By combining single-nucleus transcriptome datasets from the rhesus macaque spinal cord injury (SCI) model with developmental human spinal cord datasets, we revealed that ciliated ependymal cells respond minimally to injury and cannot revert to a developmental progenitor state. Intriguingly, we observed astrocytes transdifferentiating into mature oligodendrocytes postinjury through lineage tracing experiments. Further analysis identifies an intermediate-state glial cell population expressing both astrocyte and oligodendrocyte feature genes in adult spinal cords. The transition ratio from astrocytes into oligodendrocytes increased after remodeling injury microenvironment by functional scaffolds. Overall, our results highlight the remarkable multilineage potential of astrocytes in the adult spinal cord.
Keywords: astrocyte; lineage tracing; single-nucleus RNA sequencing; spinal cord injury; transdifferentiation.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures




References
MeSH terms
Grants and funding
- 32330059/MOST | National Natural Science Foundation of China (NSFC)
- 32071338/MOST | National Natural Science Foundation of China (NSFC)
- 32372138/MOST | National Natural Science Foundation of China (NSFC)
- 31970640/MOST | National Natural Science Foundation of China (NSFC)
- 2023YFC2412503/MOST | National Key Research and Development Program of China (NKPs)
- 2023YFC2412504/MOST | National Key Research and Development Program of China (NKPs)
- Grant No. YSBR073/CAS Project for Young Scientists in Basic Research
- 2022-I2M-3-002/CAMS Innovation Fund for Medical Sciences
- L244053/Natural Science Foundation of Beijing Municipality (Beijing Natural Science Foundation)
LinkOut - more resources
Full Text Sources
Medical

