Spatial transcriptomics unveils estrogen-modulated immune responses and structural alterations in the ectocervical mucosa of depot medroxyprogesterone acetate users
- PMID: 39762272
- PMCID: PMC11704007
- DOI: 10.1038/s41598-024-83775-9
Spatial transcriptomics unveils estrogen-modulated immune responses and structural alterations in the ectocervical mucosa of depot medroxyprogesterone acetate users
Abstract
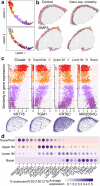
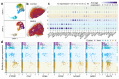
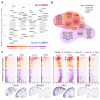
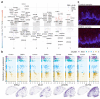
The injectable contraceptive, depot medroxyprogesterone acetate (DMPA), is associated with compromised cervical mucosal barriers. High-resolution spatial transcriptomics is applied here to reveal the spatial localization of these altered molecular markers. Ectocervical tissue samples from Kenyan sex workers using DMPA, or non-hormonal contraceptives, underwent spatial transcriptomics and gene set enrichment analyses. Integrated systemic estradiol levels and bulk tissue gene expression data from a larger cohort enhanced the study's scope. Unsupervised clustering unveiled four epithelial and seven submucosal layers, showcasing spatially restricted and diverse functional epithelial responses, and a less structured submucosal spatial ordering. DMPA associated with mucosal-wide immunoglobulin gene upregulation, verified by CD20+ B-cell immunostaining, and upregulated immune markers adjacent to the basal membrane. Downregulated genes represented spatially restricted disrupted epithelial barrier integrity and submucosal extracellular matrix dysfunction. The transcriptional profile was associated with markers of estrogen regulation. Collectively, our findings reveal estrogen-modulated distinct ectocervical transcriptional profiles associated with DMPA usage. While upregulation of immunoglobulin genes occurs throughout the mucosa, activation of innate immune responses and dysregulation of barrier integrity markers are spatially restricted. These results extend previous analyses using bulk transcriptomics and provide insights into the molecular landscape influenced by DMPA, shedding light on contraceptive effects and health implications.
Keywords: DMPA; Ectocervix; Estrogen; Hypoestrogenemia; Mucosa; Spatial transcriptomics.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests, except for Paulo Czarnewski who currently works at Precisium AI as Chief Executive Officer.
Figures


References
-
- United Nation Department of Economic and Social Affairs. 25. Contraceptive use by method 2019: data booklet. Contraception Use by Method (2019).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

