NPC1 controls TGFBR1 stability in a cholesterol transport-independent manner and promotes hepatocellular carcinoma progression
- PMID: 39762312
- PMCID: PMC11704005
- DOI: 10.1038/s41467-024-55788-5
NPC1 controls TGFBR1 stability in a cholesterol transport-independent manner and promotes hepatocellular carcinoma progression
Abstract
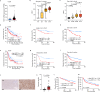
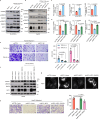
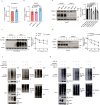
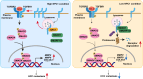
Niemann-Pick disease type C protein 1 (NPC1), classically associated with cholesterol transport and viral entry, has an emerging role in cancer biology. Here, we demonstrate that knockout of Npc1 in hepatocytes attenuates hepatocellular carcinoma (HCC) progression in both DEN (diethylnitrosamine)-CCl4 induced and MYC-driven HCC mouse models. Mechanistically, NPC1 significantly promotes HCC progression by modulating the TGF-β pathway, independent of its traditional role in cholesterol transport. We identify that the 692-854 amino acid region of NPC1's transmembrane domain is critical for its interaction with TGF-β receptor type-1 (TGFBR1). This interaction prevents the binding of SMAD7 and SMAD ubiquitylation regulatory factors (SMURFs) to TGFBR1, reducing TGFBR1 ubiquitylation and degradation, thus enhancing its stability. Notably, the NPC1 (P691S) mutant, which is defective in cholesterol transport, still binds TGFBR1, underscoring a cholesterol-independent mechanism. These findings highlight a cholesterol transport-independent mechanism by which NPC1 contributes to the stability of TGFBR1 in HCC and suggest potential therapeutic strategies targeting NPC1 for HCC treatment.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
-
- Llovet, J. M. et al. Hepatocellular carcinoma. Nat. Rev. Dis. Prim.7, 6 (2021). - PubMed
-
- Villanueva, A. Hepatocellular carcinoma. Reply. N. Engl. J. Med381, e2 (2019). - PubMed
-
- Vogel, A., Meyer, T., Sapisochin, G., Salem, R. & Saborowski, A. Hepatocellular carcinoma. Lancet400, 1345–1362 (2022). - PubMed
-
- Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin.71, 209–249 (2021). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

