The identification of a SARs-CoV2 S2 protein derived peptide with super-antigen-like stimulatory properties on T-cells
- PMID: 39762551
- PMCID: PMC11704208
- DOI: 10.1038/s42003-024-07350-8
The identification of a SARs-CoV2 S2 protein derived peptide with super-antigen-like stimulatory properties on T-cells
Abstract
Severe COVID-19 can trigger a cytokine storm, leading to acute respiratory distress syndrome (ARDS) with similarities to superantigen-induced toxic shock syndrome. An outstanding question is whether SARS-CoV-2 protein sequences can directly induce inflammatory responses. In this study, we identify a region in the SARS-CoV-2 S2 spike protein with sequence homology to bacterial super-antigens (termed P3). Computational modeling predicts P3 binding to sites on MHC class I/II and the TCR that partially overlap with sites for the binding of staphylococcal enterotoxins B and H. Like SEB and SEH derived peptides, P3 stimulated 25-40% of human CD4+ and CD8 + T-cells, increasing IFN-γ and granzyme B production. viSNE and SPADE profiling identified overlapping and distinct IFN-γ+ and GZMB+ subsets. The super-antigenic properties of P3 were further evident by its selective expansion of T-cells expressing specific TCR Vα and Vβ chain repertoires. In vivo experiments in mice revealed that the administration of P3 led to a significant upregulation of proinflammatory cytokines IL-1β, IL-6, and TNF-α. While the clinical significance of P3 in COVID-19 remains unclear, its homology to other mammalian proteins suggests a potential role for this peptide family in human inflammation and autoimmunity.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures

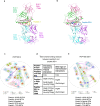

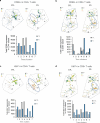


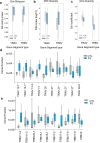
Update of
-
The identification of a SARs-CoV2 S2 protein-derived peptide with super-antigen-like stimulatory properties on T-cells.bioRxiv [Preprint]. 2024 Dec 4:2024.11.26.624714. doi: 10.1101/2024.11.26.624714. bioRxiv. 2024. Update in: Commun Biol. 2025 Jan 6;8(1):14. doi: 10.1038/s42003-024-07350-8. PMID: 39651306 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

