Identification of macrophage polarisation and mitochondria-related biomarkers in diabetic retinopathy
- PMID: 39762849
- PMCID: PMC11706200
- DOI: 10.1186/s12967-024-06038-1
Identification of macrophage polarisation and mitochondria-related biomarkers in diabetic retinopathy
Abstract
Background: The activation of macrophages or microglia in patients' whole body or local eyes play significant roles in diabetic retinopathy (DR). Mitochondrial function regulates the inflammatory polarization of macrophages. Therefore, the common mechanism of mitochondrial related genes (MRGs) and macrophage polarisation related genes (MPRGs) in DR is explored in our study to illustrate the pathophysiology of DR.
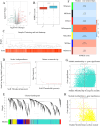
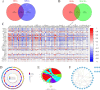
Methods: In this study, using common transcriptome data, differentially expressed genes (DEGs) were firstly analysed for GSE221521, while module genes related to MPRGs were obtained by weighted gene co-expression network analysis (WGCNA), intersections of DEGs with MRGs were taken, intersections of DEGs with module genes of the MPRGs were taken. After that, correlation analyses were performed to obtain candidate genes. Key genes were obtained by Mendelian randomisation (MR) analysis, then biomarkers were obtained by machine learning combined with receiver operating characteristic (ROC) and expression validation between DR and control cohorts in GSE221521 and GSE160306 to obtain biomarkers. Finally, biomarkers were subjected to immune infiltration analysis, gene set enrichment analysis (GSEA), and gene-gene interaction (GGI) analysis.
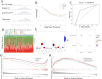
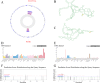
Results: A number of 784 of DEGs were taken to intersect with 1136 MRGs and 782 MPRGs, respectively, after which 89 genes with correlation were taken as candidate genes. MR analysis yielded 13 key genes with clear causal links to DR. The expression trends of PTAR1 and SLC25A34 were consistent and notable between DR cohort and control cohort in GSE221521 and GSE160306. So PTAR1 and SLC25A34 were used as biomarkers. Immune infiltration analysis showed that activated NK cell and Monocyte were notably different between DR cohort and control cohorts, and PTAR1 showed the strongest positive correlations with activated NK cell. Both biomarkers were enriched in lysosome and insulin signaling pathway. The GGI network showed that biomarkers associated with prenyltransferase activity and prenylation function.
Conclusion: This study identified two biomarkers (PTAR1 and SLC25A34) which explore the pathogenesis of DR and provide reference targets for drug development.
Keywords: Diabetic retinopathy; Macrophage polarisation; Mitochondria; PTAR1; SLC25A34.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Specific ethical approval was not required for this study because all data were obtained from sources available to the public. Consent for publication: Not appliable. Competing interests: The authors declare that they have no competing interests.
Figures

References
-
- Gangwani RA, Lian JX, McGhee SM, Wong D, Li KK. Diabetic retinopathy screening: global and local perspective. Hong Kong Med J. 2016;22:486–95. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

