Resolving the source of branch length variation in the Y chromosome phylogeny
- PMID: 39762943
- PMCID: PMC11702058
- DOI: 10.1186/s13059-024-03468-4
Resolving the source of branch length variation in the Y chromosome phylogeny
Abstract
Background: Genetic variation in the non-recombining part of the human Y chromosome has provided important insight into the paternal history of human populations. However, a significant and yet unexplained branch length variation of Y chromosome lineages has been observed, notably amongst those that are highly diverged from the human reference Y chromosome. Understanding the origin of this variation, which has previously been attributed to changes in generation time, mutation rate, or efficacy of selection, is important for accurately reconstructing human evolutionary and demographic history.
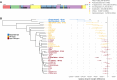
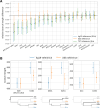
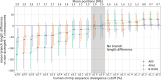
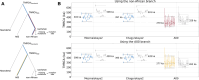
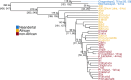
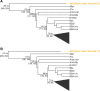
Results: Here, we analyze Y chromosomes from present-day and ancient modern humans, as well as Neandertals, and show that branch length variation amongst human Y chromosomes cannot solely be explained by differences in demographic or biological processes. Instead, reference bias results in mutations being missed on Y chromosomes that are highly diverged from the reference used for alignment. We show that masking fast-evolving, highly divergent regions of the human Y chromosome mitigates the effect of this bias and enables more accurate determination of branch lengths in the Y chromosome phylogeny.
Conclusion: We show that our approach allows us to estimate the age of ancient samples from Y chromosome sequence data and provide updated estimates for the time to the most recent common ancestor using the portion of the Y chromosome where the effect of reference bias is minimized.
Keywords: Ancient DNA; Generation time; Molecular dating; Mutation rate; Reference bias; Sequence alignment; Y chromosome.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: All sequencing data used in this study were previously published. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures

References
-
- Scozzari R, Massaia A, Trombetta B, Bellusci G, Myres NM, Novelletto A, et al. An Unbiased Resource of Novel SNP Markers Provides a New Chronology for the Human Y Chromosome and Reveals a Deep Phylogenetic Structure in Africa. Genome Res. 2014;24(3):535–44. 10.1101/gr.160788.113. - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

