This is a preprint.
The PRMT5-splicing axis is a critical oncogenic vulnerability that regulates detained intron splicing
- PMID: 39763796
- PMCID: PMC11702595
- DOI: 10.1101/2024.12.17.628905
The PRMT5-splicing axis is a critical oncogenic vulnerability that regulates detained intron splicing
Update in
-
The PRMT5-splicing axis is a critical oncogenic vulnerability that regulates detained intron splicing.iScience. 2025 Jun 20;28(7):112965. doi: 10.1016/j.isci.2025.112965. eCollection 2025 Jul 18. iScience. 2025. PMID: 40687829 Free PMC article.
Abstract
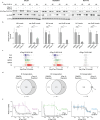
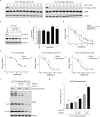
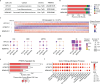
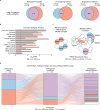
Protein arginine methyltransferase 5 (PRMT5) is a promising cancer target, yet it's unclear which PRMT5 roles underlie this vulnerability. Here, we establish that PRMT5 inhibition induces a special class of unspliced introns, called detained introns (DIs). To interrogate the impact of DIs, we depleted CLNS1A, a PRMT5 cofactor that specifically enables Sm protein methylation. We found that many, but not all, cell lines are CLNS1A-dependent and established that loss of viability is linked to loss of Sm protein methylation and DI upregulation. Finally, we discovered that PRMT5-regulated DIs, and the impacted genes, are highly conserved across human, and also mouse, cell lines but display little interspecies conservation. Despite this, human and mouse DIs have convergent impacts on proliferation by affecting essential components of proliferation-regulating complexes. Together, these data argue that the PRMT5-splicing axis, including appropriate DI splicing, underlies cancer's vulnerability to PRMT5 inhibitors.
Keywords: CLNS1A; PRMT5; detained introns; methylosome; splicing.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures



References
-
- Braun C.J., Stanciu M., Boutz P.L., Patterson J.C., Calligaris D., Higuchi F., Neupane R., Fenoglio S., Cahill D.P., Wakimoto H., et al. (2017). Coordinated Splicing of Regulatory Detained Introns within Oncogenic Transcripts Creates an Exploitable Vulnerability in Malignant Glioma. Cancer Cell 32, 411 426.e11. 10.1016/j.ccell.2017.08.018. - DOI - PMC - PubMed
-
- Engstrom L.D., Aranda R., Waters L., Moya K., Bowcut V., Vegar L., Trinh D., Hebbert A., Smith C.R., Kulyk S., et al. (2023). MRTX1719 is an MTA-cooperative PRMT5 inhibitor that exhibits synthetic lethality in preclinical models and patients with MTAP deleted cancer. Cancer Discov. 13, 2412–2431. 10.1158/2159-8290.cd-23-0669. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
