This is a preprint.
Dendritic cells type 1 control the formation, maintenance, and function of tertiary lymphoid structures in cancer
- PMID: 39763802
- PMCID: PMC11703156
- DOI: 10.1101/2024.12.27.628014
Dendritic cells type 1 control the formation, maintenance, and function of tertiary lymphoid structures in cancer
Abstract
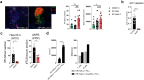
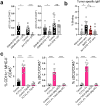
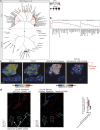
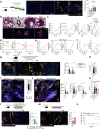
Tertiary lymphoid structures (TLS) are organized immune cell aggregates that arise in chronic inflammatory conditions. In cancer, TLS are associated with better prognosis and enhanced response to immunotherapy, making these structures attractive therapeutic targets. However, the mechanisms regulating TLS formation and maintenance in cancer are incompletely understood. Using spatial transcriptomics and multiplex imaging across various human tumors, we found an enrichment of mature dendritic cells (DC) expressing high levels of CCR7 in TLS, prompting us to investigate the role of DC in the formation and maintenance of TLS in solid tumors. To address this, we developed a novel murine model of non-small cell lung cancer (NSCLC) that forms mature TLS, containing B cell follicles with germinal centers and T cell zones with T follicular helper cells (TFH) and TCF1+PD-1+ progenitor exhausted CD8+ T cells (Tpex). Here we show that, during the early stages of tumor development, TLS formation relies on IFNγ-driven maturation of the conventional DC type 1 (cDC1) subset, their migration to tumor-draining lymph nodes (tdLN), and recruitment of activated T cells to the tumor site. As tumors progress, TLS maintenance becomes independent of T cell egress from tdLN, coinciding with a significant reduction of cDC1 migration to tdLN. Instead, mature cDC1 accumulate within intratumoral CCR7 ligand-enriched stromal hubs. Notably, timed depletion of cDC1 or disruption of their migration to these stromal hubs after TLS are formed alters TLS maintenance. Importantly, we found that cDC1-mediated antigen presentation to both CD4+ and CD8+ T cells and intact CD40 signaling, is critical for the maintenance of TLS, the preservation of the TFH cell pool, the formation of germinal center and the production of tumor-specific IgG antibodies. These findings underscore the key role of mature cDC1 in establishing and maintaining functional TLS within tumor lesions and highlight the potential for cDC1-targeting therapies as a promising strategy to enhance TLS function and improve anti-tumor immunity in patients with cancer.
Figures






References
-
- Alspach E., Lussier D.M., Miceli A.P., Kizhvatov I., DuPage M., Luoma A.M., Meng W., Lichti C.F., Esaulova E., Vomund A.N., Runci D., Ward J.P., Gubin M.M., Medrano R.F.V., Arthur C.D., White J.M., Sheehan K.C.F., Chen A., Wucherpfennig K.W., Jacks T., Unanue E.R., Artyomov M.N., Schreiber R.D., 2019. MHC-II neoantigens shape tumour immunity and response to immunotherapy. Nature 574, 696–701. 10.1038/s41586-019-1671-8 - DOI - PMC - PubMed
-
- Belabed M., Park M.D., Blouin C.M., Balan S., Moon C.Y., Boumelha J., Peros A., Mattiuz R., Reid A.M., Azimi C.S., LaMarche N.M., Troncoso L., Amabile A., Berichel J.L., Chen S.T., Wilk C.M., Brown B.D., Radford K., Ghosh S., Rothlin C.V., Yvan-Charvet L., Marron T.U., Puleston D.J., Bhardwaj N., Lamaze C., Merad M., 2023. AXL limits the mobilization of cholesterol to regulate dendritic cell maturation and the immunogenic response to cancer. 10.1101/2023.12.25.573303 - DOI - PubMed
-
- Cabrita R., Lauss M., Sanna A., Donia M., Skaarup Larsen M., Mitra S., Johansson I., Phung B., Harbst K., Vallon-Christersson J., van Schoiack A., Lövgren K., Warren S., Jirström K., Olsson H., Pietras K., Ingvar C., Isaksson K., Schadendorf D., Schmidt H., Bastholt L., Carneiro A., Wargo J.A., Svane I.M., Jönsson G., 2020. Tertiary lymphoid structures improve immunotherapy and survival in melanoma. Nature 577, 561–565. 10.1038/s41586-019-1914-8 - DOI - PubMed
-
- Chaurio R.A., Anadon C.M., Lee Costich T., Payne K.K., Biswas S., Harro C.M., Moran C., Ortiz A.C., Cortina C., Rigolizzo K.E., Sprenger K.B., Mine J.A., Innamarato P., Mandal G., Powers J.J., Martin A., Wang Z., Mehta S., Perez B.A., Li R., Robinson J., Kroeger J.L., Curiel T.J., Yu X., Rodriguez P.C., Conejo-Garcia J.R., 2022. TGF-β-mediated silencing of genomic organizer SATB1 promotes Tfh cell differentiation and formation of intra-tumoral tertiary lymphoid structures. Immunity 55, 115–128.e9. 10.1016/j.immuni.2021.12.007 - DOI - PMC - PubMed
-
- Cui C., Wang J., Fagerberg E., Chen P.-M., Connolly K.A., Damo M., Cheung J.F., Mao T., Askari A.S., Chen S., Fitzgerald B., Foster G.G., Eisenbarth S.C., Zhao H., Craft J., Joshi N.S., 2021. Neoantigen-driven B cell and CD4 T follicular helper cell collaboration promotes anti-tumor CD8 T cell responses. Cell 184, 6101–6118.e13. 10.1016/j.cell.2021.11.007 - DOI - PMC - PubMed
Publication types
Grants and funding
- U24 CA224319/CA/NCI NIH HHS/United States
- R01 DA055434/DA/NIDA NIH HHS/United States
- R33 CA263705/CA/NCI NIH HHS/United States
- K00 CA223043/CA/NCI NIH HHS/United States
- R01 AI153363/AI/NIAID NIH HHS/United States
- R01 DK122853/DK/NIDDK NIH HHS/United States
- R37 CA230636/CA/NCI NIH HHS/United States
- U2C ES030859/ES/NIEHS NIH HHS/United States
- R01 CA257195/CA/NCI NIH HHS/United States
- U01 OH012621/OH/NIOSH CDC HHS/United States
- R01 CA154947/CA/NCI NIH HHS/United States
- U01 DK124165/DK/NIDDK NIH HHS/United States
- R01 CA251155/CA/NCI NIH HHS/United States
- R01 HL166720/HL/NHLBI NIH HHS/United States
- R01 CA254104/CA/NCI NIH HHS/United States
- P30 CA196521/CA/NCI NIH HHS/United States
- R01 DK122253/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
