This is a preprint.
PinkyCaMP a mScarlet-based calcium sensor with exceptional brightness, photostability, and multiplexing capabilities
- PMID: 39763884
- PMCID: PMC11702558
- DOI: 10.1101/2024.12.16.628673
PinkyCaMP a mScarlet-based calcium sensor with exceptional brightness, photostability, and multiplexing capabilities
Abstract
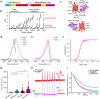
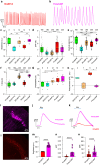
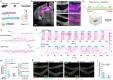
Genetically encoded calcium (Ca2+) indicators (GECIs) are widely used for imaging neuronal activity, yet current limitations of existing red fluorescent GECIs have constrained their applicability. The inherently dim fluorescence and low signal-to-noise ratio of red-shifted GECIs have posed significant challenges. More critically, several red-fluorescent GECIs exhibit photoswitching when exposed to blue light, thereby limiting their applicability in all-optical experimental approaches. Here, we present the development of PinkyCaMP, the first mScarlet-based Ca2+ sensor that outperforms current red fluorescent sensors in brightness, photostability, signal-to-noise ratio, and compatibility with optogenetics and neurotransmitter imaging. PinkyCaMP is well-tolerated by neurons, showing no toxicity or aggregation, both in vitro and in vivo. All imaging approaches, including single-photon excitation methods such as fiber photometry, widefield imaging, miniscope imaging, as well as two-photon imaging in awake mice, are fully compatible with PinkyCaMP.
Conflict of interest statement
Conflict of interest No conflict of interest.
Figures



References
-
- Tian L., Hires S. A. & Looger L. L. Imaging neuronal activity with genetically encoded calcium indicators. Cold Spring Harb. Protoc. 2012, 647–656 (2012). - PubMed
-
- Nakai J., Ohkura M. & Imoto K. A high signal-to-noise Ca(2+) probe composed of a single green fluorescent protein. Nat. Biotechnol. 19, 137–141 (2001). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
