This is a preprint.
The pseudoknot structure of a viral RNA reveals a conserved mechanism for programmed exoribonuclease resistance
- PMID: 39763890
- PMCID: PMC11702639
- DOI: 10.1101/2024.12.17.628992
The pseudoknot structure of a viral RNA reveals a conserved mechanism for programmed exoribonuclease resistance
Abstract
Exoribonuclease-resistant RNAs (xrRNAs) are viral RNA structures that block degradation by cellular 5'-3' exoribonucleases to produce subgenomic viral RNAs during infection. Initially discovered in flaviviruses, xrRNAs have since been identified in wide range of RNA viruses, including those that infect plants. High sequence variability among viral xrRNAs raises questions about the shared molecular features that characterize this functional RNA class. Here, we present the first structure of a plant-virus xrRNA in its active exoribonuclease-resistant conformation. The xrRNA forms a 9 base pair pseudoknot that creates a knot-like topology similar to that of flavivirus xrRNAs, despite lacking sequence similarity. Biophysical assays confirm a compact pseudoknot structure in solution, and functional studies validate its relevance both in vitro and during infection. Our study reveals how viral RNAs achieve a common functional outcome through highly divergent sequences and identifies the knot-like topology as a defining feature of xrRNAs.
Figures



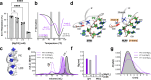

References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
