This is a preprint.
Eukaryotic composition across seasons and social groups in the gut microbiota of wild baboons
- PMID: 39763902
- PMCID: PMC11702614
- DOI: 10.1101/2024.12.17.628920
Eukaryotic composition across seasons and social groups in the gut microbiota of wild baboons
Update in
-
Eukaryotic composition across seasons and social groups in the gut microbiota of wild baboons.Anim Microbiome. 2025 Jun 21;7(1):70. doi: 10.1186/s42523-025-00436-6. Anim Microbiome. 2025. PMID: 40544290 Free PMC article.
Abstract
Background: Animals coexist with complex microbiota, including bacteria, viruses, and eukaryotes (e.g., fungi, protists, and helminths). While the composition of bacterial and viral components of animal microbiota are increasingly well understood, eukaryotic composition remains neglected. Here we characterized eukaryotic diversity in the microbiomes in wild baboons and tested the degree to which eukaryotic community composition was predicted by host social group membership, sex, age, and season of sample collection.
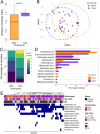
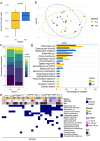
Results: We analyzed a total of 75 fecal samples collected between 2012 and 2014 from 73 wild baboons in the Amboseli ecosystem in Kenya. DNA from these samples was subjected to shotgun metagenomic sequencing, revealing members of the kingdoms Protista, Chromista, and Fungi in 90.7%, 46.7%, and 20.3% of samples, respectively. Social group membership explained 11.2% of the global diversity in gut eukaryotic species composition, but we did not detect statistically significant effect of season, host age, and host sex. Across samples, the most prevalent protists were Entamoeba coli (74.66% of samples), Enteromonas hominis (53.33% of samples), and Blastocystis subtype 3 (38.66% of samples), while the most prevalent fungi included Pichia manshurica (14.66% of samples), and Ogataea naganishii (6.66% of samples).
Conclusions: Protista, Chromista, and Fungi are common members of the gut microbiome of wild baboons. More work on eukaryotic members of primate gut microbiota is essential for primate health monitoring and management strategies.
Keywords: eukaryotes; fungi; gut microbiome; protists; social groups; wild baboons.
Figures
References
-
- Mas-Lloret J, Obón-Santacana M, Ibáñez-Sanz G, Guinó E, Pato ML, Rodriguez-Moranta F, et al. Gut microbiome diversity detected by high-coverage 16S and shotgun sequencing of paired stool and colon sample. Scientific Data [Internet]. 2020;7. Available from: 10.1038/s41597-020-0427-5 - DOI - PMC - PubMed
-
- Wei F, Wu Q, Hu Y, Huang G, Nie Y, Yan L. Conservation metagenomics: a new branch of conservation biology. Sci China Life Sci. 2019; 62:168–78. - PubMed
-
- Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, et al. Host-gut microbiota metabolic interactions. Science. 2012; 336:1262–7. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
