This is a preprint.
Comparative genomics of the sexually transmitted parasite Trichomonas vaginalis reveals relaxed and convergent evolution and genes involved in spillover from birds to humans
- PMID: 39763951
- PMCID: PMC11703204
- DOI: 10.1101/2024.12.22.629724
Comparative genomics of the sexually transmitted parasite Trichomonas vaginalis reveals relaxed and convergent evolution and genes involved in spillover from birds to humans
Update in
-
Comparative genomics of the parasite Trichomonas vaginalis reveals genes involved in spillover from birds to humans.Nat Commun. 2025 Jul 24;16(1):6487. doi: 10.1038/s41467-025-61483-w. Nat Commun. 2025. PMID: 40707449 Free PMC article.
Abstract
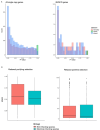
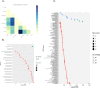
Trichomonas vaginalis is the causative agent of the venereal disease trichomoniasis which infects men and women globally and is associated with serious outcomes during pregnancy and cancers of the human reproductive tract. Trichomonads parasitize a range of hosts in addition to humans including birds, livestock, and domesticated animals. Recent genetic analysis of trichomonads recovered from columbid birds has provided evidence that these parasite species undergo frequent host-switching, and that a current epoch spillover event from columbids likely gave rise to T. vaginalis in humans. We undertook a comparative evolutionary genomics study of seven trichomonad species, generating chromosome-scale reference genomes for T. vaginalis and its avian sister species Trichomonas stableri, and assemblies of five other species that infect birds and mammals. Human-infecting trichomonad lineages have undergone recent and convergent genome size expansions compared to their avian sister species, and the major contributor to their increased genome size is increased repeat expansions, especially multicopy gene families and transposable elements, with genetic drift likely a driver due to relaxed selection. Trichomonads have independently host-switched twice from birds to humans, and genes implicated in the transition to the human host include those associated with host tissue adherence and phagocytosis, extracellular vesicles, and CAZyme virulence factors.
Conflict of interest statement
Competing interests The authors declare no competing interests.
Figures

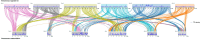



References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
