This is a preprint.
Deep learning-based prediction of chemical accumulation in a pathogenic mycobacterium
- PMID: 39764009
- PMCID: PMC11702553
- DOI: 10.1101/2024.12.15.628588
Deep learning-based prediction of chemical accumulation in a pathogenic mycobacterium
Abstract
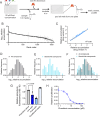
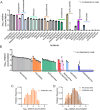
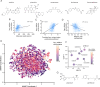
Drugs must accumulate at their target site to be effective, and inadequate uptake of drugs is a substantial barrier to the design of potent therapies. This is particularly true in the development of antibiotics, as bacteria possess numerous barriers to prevent chemical uptake. Designing compounds that circumvent bacterial barriers and accumulate to high levels in cells could dramatically improve the success rate of antibiotic candidates. However, a comprehensive understanding of which chemical structures promote or prevent drug uptake is currently lacking. Here we use liquid chromatography-mass spectrometry to measure accumulation of 1528 approved drugs in Mycobacterium abscessus, a highly drug-resistant, opportunistic pathogen. We find that simple chemical properties fail to effectively predict drug accumulation in mycobacteria. Instead, we use our data to train deep learning models that predict drug accumulation in M. abscessus with high accuracy, including for chemically diverse compounds not included in our original drug library. We find that differential drug uptake is a critical determinant of the efficacy of drugs currently in development and can identify compounds which accumulate well and have antibacterial activity in M. abscessus. These predictive algorithms can be an important complement to chemical synthesis and accumulation assays in the evaluation of drug candidates.
Keywords: Drug uptake; chemical permeability; deep learning; mycobacteria.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures


Similar articles
-
Efflux pumps and membrane permeability contribute to intrinsic antibiotic resistance in Mycobacterium abscessus.bioRxiv [Preprint]. 2024 Aug 24:2024.08.23.609441. doi: 10.1101/2024.08.23.609441. bioRxiv. 2024. Update in: PLoS Pathog. 2025 Apr 10;21(4):e1013027. doi: 10.1371/journal.ppat.1013027. PMID: 39229117 Free PMC article. Updated. Preprint.
-
The Role of Antibiotic-Target-Modifying and Antibiotic-Modifying Enzymes in Mycobacterium abscessus Drug Resistance.Front Microbiol. 2018 Sep 12;9:2179. doi: 10.3389/fmicb.2018.02179. eCollection 2018. Front Microbiol. 2018. PMID: 30258428 Free PMC article. Review.
-
Genome-Wide Essentiality Analysis of Mycobacterium abscessus by Saturated Transposon Mutagenesis and Deep Sequencing.mBio. 2021 Jun 29;12(3):e0104921. doi: 10.1128/mBio.01049-21. Epub 2021 Jun 15. mBio. 2021. PMID: 34126767 Free PMC article.
-
Noncanonical Mismatch Repair Protein NucS Modulates the Emergence of Antibiotic Resistance in Mycobacterium abscessus.Microbiol Spectr. 2022 Dec 21;10(6):e0222822. doi: 10.1128/spectrum.02228-22. Epub 2022 Oct 11. Microbiol Spectr. 2022. PMID: 36219122 Free PMC article.
-
A laboratory perspective on Mycobacterium abscessus biofilm culture, characterization and drug activity testing.Front Microbiol. 2024 Apr 16;15:1392606. doi: 10.3389/fmicb.2024.1392606. eCollection 2024. Front Microbiol. 2024. PMID: 38690364 Free PMC article. Review.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
