This is a preprint.
Mediator kinase inhibition drives myometrial stem cell differentiation and the uterine fibroid phenotype through super-enhancer reprogramming
- PMID: 39764110
- PMCID: PMC11702794
- DOI: 10.21203/rs.3.rs-5125876/v1
Mediator kinase inhibition drives myometrial stem cell differentiation and the uterine fibroid phenotype through super-enhancer reprogramming
Update in
-
Mediator kinase inhibition drives myometrial stem cell differentiation and the uterine fibroid phenotype through super-enhancer reprogramming.J Mol Med (Berl). 2025 Mar;103(3):311-326. doi: 10.1007/s00109-025-02517-0. Epub 2025 Feb 4. J Mol Med (Berl). 2025. PMID: 39904883 Free PMC article.
Abstract
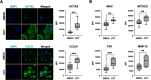
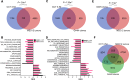
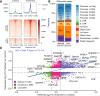
Uterine fibroids (UFs) are the most common non-cutaneous tumors in women worldwide. UFs arise from genetic alterations in myometrial stem cells (MM SCs) that trigger their transformation into tumor initiating cells (UF SCs). Mutations in the RNA polymerase II Mediator subunit MED12 are dominant drivers of UFs, accounting for 70% of these clinically significant lesions. Biochemically, UF driver mutations in MED12 disrupt CDK8/19 kinase activity in Mediator, but how Mediator kinase disruption triggers MM SC transformation remains unknown. Here, we show that pharmacologic inhibition of CDK8/19 in MM SCs removes a barrier to myogenic differentiation down an altered pathway characterized by molecular phenotypes characteristic of UFs, including oncogenic growth and extracellular matrix (ECM) production. These perturbations appear to be induced by transcriptomic changes, arising in part through epigenomic alteration and super-enhancer reprogramming, that broadly recapitulate those found in MED12-mutant UFs. Altogether these findings provide new insights concerning the biological role of CDK8/19 in MM SC biology and UF formation.
Conflict of interest statement
Competing Interests The authors have no relevant financial or non-financial interests to disclose.
Figures



References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

