This is a preprint.
Triple coding in human SRD5A1 mRNA
- PMID: 39764142
- PMCID: PMC11702784
- DOI: 10.21203/rs.3.rs-5390104/v1
Triple coding in human SRD5A1 mRNA
Abstract
Background: Nucleotide sequence can be translated in three reading frames from 5' to 3' producing distinct protein products. Many examples of RNA translation in two reading frames (dual coding) have been identified so far.
Results: We report simultaneous translation of mRNA transcripts derived from SRD5A1 locus in all three reading frames that result in the synthesis of long proteins. This occurs due to initiation at three nearby AUG codons occurring in all three-reading frame. Only one of the three proteoforms contains the conserved catalytical domain of SDRD5A1 produced either from the second or the third AUG codon depending on the transcript. Paradoxically, ribosome profiling data and expression reporters indicate that the most efficient translation produces catalytically inactive proteoforms. While phylogenetic analysis suggests that the long triple decoding region is specific to primates, occurrence of nearby AUGs in all three reading frames is ancestral to placental mammals. This suggests that their evolutionary significance belongs to regulation of translation rather than biological role of their products. By analysing multiple publicly available ribosome profiling data and with gene expression assays carried out in different cellular environments, we show that relative expression of these proteoforms is mutually dependent and vary across environments supporting this conjecture. A remarkable feature of triple decoding is its resistance to indel mutations with apparent implications to clinical interpretation of genomic variants.
Conclusion: We argue for the importance of identification, characterisation and annotation of productive RNA translation irrespective of the presumed biological roles of the products of this translation.
Keywords: Overlapping genes; SRD5A1; gene annotation; protein synthesis; ribosome decision graphs; translation control; translation initiation; translon; uORF.
Conflict of interest statement
Competing interests PVB and GL are cofounders of EIRNABio
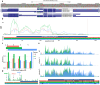
Figures


References
-
- Andreev Dmitry E., O’Connor Patrick B. F., Zhdanov Alexander V., Dmitriev Ruslan I., Shatsky Ivan N., Papkovsky Dmitri B., and Baranov Pavel V.. 2015. ‘Oxygen and Glucose Deprivation Induces Widespread Alterations in mRNA Translation within 20 Minutes’. Genome Biology 16 (1): 90. 10.1186/s13059-015-0651-z. - DOI - PMC - PubMed
-
- Anisimova Aleksandra S., Kolyupanova Natalia M., Makarova Nadezhda E., Egorov Artyom A., Kulakovskiy Ivan V., and Dmitriev Sergey E.. 2023. ‘Human Tissues Exhibit Diverse Composition of Translation Machinery’. International Journal of Molecular Sciences 24 (9): 8361. 10.3390/ijms24098361. - DOI - PMC - PubMed
-
- Atkins John F., Wills Norma M., Loughran Gary, Wu Chih-Yu, Parsawar Krishna, Ryan Martin D., Wang Chung-Hsiung, and Nelson Chad C.. 2007. ‘A Case for “StopGo”: Reprogramming Translation to Augment Codon Meaning of GGN by Promoting Unconventional Termination (Stop) after Addition of Glycine and Then Allowing Continued Translation (Go)’. RNA (New York, N.Y.) 13 (6): 803–10. 10.1261/rna.487907. - DOI - PMC - PubMed
-
- Chang Kai-Hsiung, Li Rui, Papari-Zareei Mahboubeh, Watumull Lori, Zhao Yan Daniel, Auchus Richard J., and Sharifi Nima. 2011. ‘Dihydrotestosterone Synthesis Bypasses Testosterone to Drive Castration-Resistant Prostate Cancer’. Proceedings of the National Academy of Sciences of the United States of America 108 (33): 13728–33. 10.1073/pnas.1107898108. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

