Core transcriptome network modulates temperature (heat and cold) and osmotic (drought, salinity, and waterlogging) stress responses in oil palm
- PMID: 39764238
- PMCID: PMC11701223
- DOI: 10.3389/fpls.2024.1497017
Core transcriptome network modulates temperature (heat and cold) and osmotic (drought, salinity, and waterlogging) stress responses in oil palm
Abstract
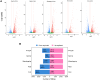
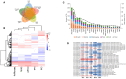
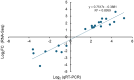
Oil palm (Elaeis guineensis) yield is impacted by abiotic stresses, leading to significant economic losses. To understand the core abiotic stress transcriptome (CAST) of oil palm, we performed RNA-Seq analyses of oil palm leaves subjected to drought, salinity, waterlogging, heat, and cold stresses. A total of 19,834 differentially expressed genes (DEGs) were identified. Cold treatment induced the highest number of DEGs (5,300), followed by heat (4,114), drought (3,751), waterlogging (3,573), and, lastly, salinity (3096) stress. Subsequent analysis revealed the CAST of oil palm, comprising 588 DEGs commonly expressed under drought, salinity, waterlogging, heat, and cold stress conditions. Function annotation of these DEGs suggests their roles in signal transduction, transcription regulation, and abiotic stress responses including synthesis of osmolytes, secondary metabolites, and molecular chaperones. Moreover, we identified core DEGs encoding kinases, ERF, NAC TFs, heat shock proteins, E3 ubiquitin-protein ligase, terpineol synthase, and cytochrome P450. These core DEGs may be potential key modulators that interplay in triggering rapid abiotic stress responses to achieve delicate equilibrium between productivity and adaptation to abiotic stresses. This comprehensive study provides insights into the key modulators in the CAST of oil palm, and their potential applications as markers for selecting climate-resilient oil palms or opportunities to develop future climate resilient oil palm using genome editing.
Keywords: RNA-seq; abiotic stresses; core transcriptome; differential expressed genes; oil palm.
Copyright © 2024 Lee, Yeap, Kee, Kulaveerasingam and Ross Appleton.
Conflict of interest statement
All authors were employed by SD Guthrie Technology Centre / SD Guthrie Research, Malaysia.
Figures






References
-
- Ali Z., Merrium S., Habib-ur-Rahman M., Hakeem S., Saddique M. A. B., Sher M. A. (2022). Wetting mechanism and morphological adaptation; leaf rolling enhancing atmospheric water acquisition in wheat crop—a review. Environ. Sci. pollut. Res. 29, 30967–30985. doi: 10.1007/s11356-022-18846-3 - DOI - PMC - PubMed
-
- Anders S., Huber W. (2012). Differential expression of RNA-Seq data at the gene level–the DESeq package. Heidelberg Germany: Eur. Mol. Biol. Lab. 10.
-
- Azzeme A. M., Abdullah S. N. A., Aziz M. A., Wahab P. E. M. (2016). Oil palm leaves and roots differ in physiological response, antioxidant enzyme activities and expression of stress-responsive genes upon exposure to drought stress. Acta Physiologiae Plantarum 38, 52. doi: 10.1007/s11738-016-2073-2 - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

