Integrated View of Baseline Protein Expression in Human Tissues Using Public Data Independent Acquisition Data Sets
- PMID: 39764611
- PMCID: PMC11811993
- DOI: 10.1021/acs.jproteome.4c00788
Integrated View of Baseline Protein Expression in Human Tissues Using Public Data Independent Acquisition Data Sets
Abstract
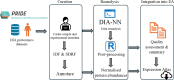
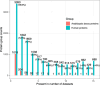
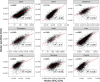
The PRIDE database is the largest public data repository of mass spectrometry-based proteomics data and currently stores more than 40,000 data sets covering a wide range of organisms, experimental techniques, and biological conditions. During the past few years, PRIDE has seen a significant increase in the amount of submitted data-independent acquisition (DIA) proteomics data sets. This provides an excellent opportunity for large-scale data reanalysis and reuse. We have reanalyzed 15 public label-free DIA data sets across various healthy human tissues to provide a state-of-the-art view of the human proteome in baseline conditions (without any perturbations). We computed baseline protein abundances and compared them across various tissues, samples, and data sets. Our second aim was to compare protein abundances obtained here from the results of previous analyses using human baseline data-dependent acquisition (DDA) data sets. We observed a good correlation across some tissues, especially in the liver and colon, but weak correlations were found in others, such as the lung and pancreas. The reanalyzed results including protein abundance values and curated metadata are made available to view and download from the resource Expression Atlas.
Keywords: Expression Atlas; PRIDE; baseline expression; data independent acquisition; data reanalysis; mass spectrometry; proteomics.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


Similar articles
-
Integrated Proteomics Analysis of Baseline Protein Expression in Pig Tissues.J Proteome Res. 2024 Jun 7;23(6):1948-1959. doi: 10.1021/acs.jproteome.3c00741. Epub 2024 May 8. J Proteome Res. 2024. PMID: 38717300 Free PMC article.
-
Integrated View of Baseline Protein Expression in Human Tissues.J Proteome Res. 2023 Mar 3;22(3):729-742. doi: 10.1021/acs.jproteome.2c00406. Epub 2022 Dec 28. J Proteome Res. 2023. PMID: 36577097 Free PMC article.
-
Implementing the reuse of public DIA proteomics datasets: from the PRIDE database to Expression Atlas.Sci Data. 2022 Jun 14;9(1):335. doi: 10.1038/s41597-022-01380-9. Sci Data. 2022. PMID: 35701420 Free PMC article.
-
Tissue proteomics repositories for data reanalysis.Mass Spectrom Rev. 2024 Nov-Dec;43(6):1270-1284. doi: 10.1002/mas.21860. Epub 2023 Aug 3. Mass Spectrom Rev. 2024. PMID: 37534389 Review.
-
Multiplexed and data-independent tandem mass spectrometry for global proteome profiling.Mass Spectrom Rev. 2014 Nov-Dec;33(6):452-70. doi: 10.1002/mas.21400. Epub 2013 Nov 26. Mass Spectrom Rev. 2014. PMID: 24281846 Review.
References
-
- Gillet L. C.; Navarro P.; Tate S.; Rost H.; Selevsek N.; Reiter L.; Bonner R.; Aebersold R. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol. Cell. Proteomics 2012, 11 (6), O111.016717.10.1074/mcp.O111.016717. - DOI - PMC - PubMed
-
- Perez-Riverol Y.; Bai J.; Bandla C.; Garcia-Seisdedos D.; Hewapathirana S.; Kamatchinathan S.; Kundu D. J.; Prakash A.; Frericks-Zipper A.; Eisenacher M.; et al. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res 2022, 50 (D1), D543–D552. 10.1093/nar/gkab1038. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

