Impact of LITAF on Mitophagy and Neuronal Damage in Epilepsy via MCL-1 Ubiquitination
- PMID: 39764629
- PMCID: PMC11705406
- DOI: 10.1111/cns.70191
Impact of LITAF on Mitophagy and Neuronal Damage in Epilepsy via MCL-1 Ubiquitination
Abstract
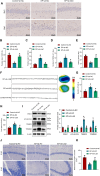
Objective: This study aims to investigate how the E3 ubiquitin ligase LITAF influences mitochondrial autophagy by modulating MCL-1 ubiquitination, and its role in the development of epilepsy.
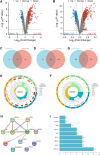
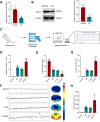
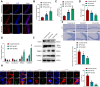
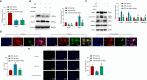
Methods: Employing single-cell RNA sequencing (scRNA-seq) to analyze brain tissue from epilepsy patients, along with high-throughput transcriptomics, we identified changes in gene expression. This was complemented by in vivo and in vitro experiments, including protein-protein interaction (PPI) network analysis, western blotting, and behavioral assessments in mouse models.
Results: Neuronal cells in epilepsy patients exhibited significant gene expression alterations, with increased activity in apoptosis-related pathways and decreased activity in neurotransmitter-related pathways. LITAF was identified as a key upregulated factor, inhibiting mitochondrial autophagy by promoting MCL-1 ubiquitination, leading to increased neuronal damage. Knockdown experiments in mouse models further confirmed that LITAF facilitates MCL-1 ubiquitination, aggravating neuronal injury.
Conclusion: Our findings demonstrate that LITAF regulates MCL-1 ubiquitination, significantly impacting mitochondrial autophagy and contributing to neuronal damage in epilepsy. Targeting LITAF and its downstream mechanisms may offer a promising therapeutic strategy for managing epilepsy.
Keywords: LPS‐induced TNF‐alpha factor; MCL1; epilepsy; mitochondrial autophagy; neuroprotection; ubiquitination regulation.
© 2025 The Author(s). CNS Neuroscience & Therapeutics published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Zhong W., Johnson C. M., Cui N., Xing H., Wu Y., and Jiang C., “Effects of Chronic Exposure to Low Dose THIP on Brainstem Neuronal Excitability in Mouse Models of Rett Syndrome: Evidence From Symptomatic Females,” Neuropharmacology 116 (2017): 288–299, 10.1016/j.neuropharm.2017.01.002. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

