Fine-tuning probes for fluorescence polarization binding assays of bivalent ligands against polo-like kinase 1 using full-length protein
- PMID: 39764864
- PMCID: PMC12349915
- DOI: 10.1016/j.bmc.2024.118055
Fine-tuning probes for fluorescence polarization binding assays of bivalent ligands against polo-like kinase 1 using full-length protein
Abstract
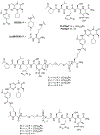
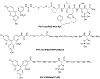
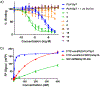
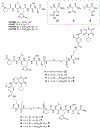
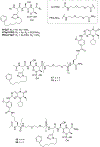
Polo-like kinase 1 (Plk1) is an important cell cycle regulator that is a recognized target for development of anti-cancer therapeutics. Plk1 is composed of a catalytic kinase domain (KD), a flexible interdomain linker and a polo-box domain (PBD). Intramolecular protein-protein interactions (PPIs) between the PBD and KD result in "auto-inhibition" that is an essential component of proper Plk1 function. Recently, we developed high-affinity PBD-binding inhibitors using a bivalent approach. These ligands contain the low-nanomolar affinity Plk1 KD-binding inhibitors BI2536 or Wortmannin tethered to the PBD-binding peptide, PLH*SpT (H* represents a -(CH2)8Ph group on the histidine side chain π-nitrogen). Due to the extremely high affinity of these bivalent inhibitors, to avoid bottoming out in competitive binding assays, it was necessary to use PLH*SpT in the affinity probe. As reported herein, we have developed fluorescence polarization assays using a new fluorescent probe based on the Plk1 PBD-binding peptide, FDPPLHSpTA. We applied the assay to evaluate the affinities of bivalent inhibitors that possess a variety of PBD-binding peptides having much lower PBD-affinities than PLH*SpT. Tethering BI2536 in these bivalent inhibitors resulted in significant affinity enhancements as compared to the parent monovalent peptides.
Keywords: Bivalent inhibitor; Fluorescence Polarization (FP) assay; Phosphorylated threonine (pT); Polo-box domain (PBD); Polo-like kinase 1 (Plk1).
Copyright © 2024 The Author(s). Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

References
-
- Pawson T, Nash P Handbook of Cell Signaling (Second Edition), eds. Bradshaw RA and Dennis EA, Academic Press, San Diego, CA: 2010: pp. 399–411. 10.1016/B978-0-12-374145-5.00057-7 - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

