An Epidemiological Study on Salmonella in Tibetan Yaks from the Qinghai-Tibet Plateau Area in China
- PMID: 39765601
- PMCID: PMC11672581
- DOI: 10.3390/ani14243697
An Epidemiological Study on Salmonella in Tibetan Yaks from the Qinghai-Tibet Plateau Area in China
Abstract
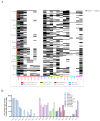
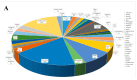
Salmonella is an important foodborne pathogen that can cause a range of illnesses in humans; it has also been a key focus for monitoring in the field of public health, including gastroenteritis, sepsis, and arthritis, and can also cause a decline in egg production in poultry and diarrhea and abortion in livestock, leading to death in severe cases, resulting in huge economic losses. This study aimed to investigate the isolation rate, antimicrobial resistance, serotypes, and genetic diversity of Salmonella isolated from yak feces in various regions on the Qinghai-Tibet Plateau. A total of 1222 samples of yak dung were collected from major cities in the Qinghai-Tibet Plateau area, and the sensitivity of the isolated bacteria to 10 major classes of antibiotics was determined using the K-B paper disk diffusion method for drug susceptibility. Meanwhile, the serotypes of the isolated bacteria were analyzed using the plate agglutination test for serum antigens, and their carriage of drug resistance and virulence genes was determined using PCR and gel electrophoresis experiments. The isolated bacteria were also classified using MLST (Multi-Locus Sequence Typing). The overall isolation rate for Salmonella was 18.25% (223/1222), and the results of the antibiotic susceptibility tests showed that 98.65% (220/223) of the isolated bacteria were resistant to multiple antibiotics. In the 223 isolates of Salmonella, eight classes of 20 different resistance genes, 30 serotypes, and 15 different types of virulence genes were detected. The MLST analysis identified 45 distinct sequence types (STs), including five clonal complexes, of which ST34, ST11, and ST19 were the most common. These findings contribute valuable information about strain resources, genetic profiles, and typing data for Salmonella in the Qinghai-Tibet Plateau area, facilitating improved bacterial surveillance, identification, and control in yak populations. They also provide certain data supplements for animal Salmonella infections globally, filling research gaps.
Keywords: Salmonella; drug resistance; multi-locus sequence typing; serotype; yaks.
Conflict of interest statement
The authors declare that this research was conducted in the absence of any commercial or financial relationships that could be construed as potential conflicts of interest.
Figures









References
-
- Andreychev A., Boyarova E., Brandler O., Tukhbatullin A., Kapustina S. Terrestrial and subterranean mammals as reservoirsof zoonotic diseases in the central part of European Russia. Diversity. 2023;15:39. doi: 10.3390/d15010039. - DOI
LinkOut - more resources
Full Text Sources

