Genome-Wide Microsatellites in Acanthopagrus latus: Development, Distribution, Characterization, and Polymorphism
- PMID: 39765613
- PMCID: PMC11672618
- DOI: 10.3390/ani14243709
Genome-Wide Microsatellites in Acanthopagrus latus: Development, Distribution, Characterization, and Polymorphism
Abstract
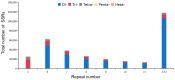
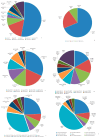
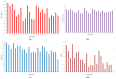
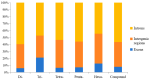
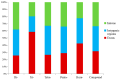
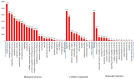
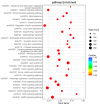
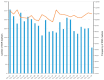
The yellowfin seabream (Acanthopagrus latus) is an economically important commercial mariculture fish in China and Southeast Asia. Only a few simple sequence repeats (SSRs) of A. latus have been isolated and reported, which has hindered breeding progress. A total of 318,862 SSRs were isolated and characterized from the A. latus genome in this study. All SSRs were 9,069,670 bp in length, accounting for 1.32% of the genome. The density and frequency of SSRs were 468.40 loci/Mb and 13,323.19 bp/Mb, respectively. The major SSRs were dinucleotides (accounting for 76.92%), followed by trinucleotides (15.75%). The most abundant SSR motif was (AC)n (168,390, accounting for 53%), with the highest frequency (245.78 loci/Mb) and density (7304.18 bp/Mb). Most SSRs were located in non-coding regions, such as intergenic regions (34.54%) and introns (56.91%). SSR-containing exons were distributed into 51 gene ontology (GO) terms and significantly enriched in immunity- and growth-related pathways. A total of 217,791 SSR markers were successfully designed. Nine SSR markers were amplified in 29 A. latus individuals, and eight of them possess high polymorphism. The cross-species transferability of 33 out of the 37 tested loci were successfully amplified in Acanthopagrus schlegelii. These results lay the foundation for the molecular marker-assisted breeding and genetic information assessment of A. latus.
Keywords: Acanthopagrus latus; GO; KEGG; genome-wide; microsatellite.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
References
-
- Morshedi V., Hamedi S., Pourkhazaei F., Torfi Mozanzadeh M., Tamadoni R., Ebadi M., Esmaili A., Azodi M., Gisbert E. Larval rearing and ontogeny of digestive enzyme activities in yellowfin seabream (Acanthopagrus latus, Houttuyn 1782) Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2021;261:111044. doi: 10.1016/j.cbpa.2021.111044. - DOI - PubMed
-
- Bureau of Fisheries. Ministry of Agriculture of China . China Fishery Statistical Yearbook. China Statistics Press; Beijing, China: 2024. p. 22.
-
- Zhu K.C., Zhang N., Liu B.S., Guo L., Guo H.Y., Jiang S.G., Zhang D.C. A chromosome-level genome assembly of the yellowfin seabream (Acanthopagrus latus; Hottuyn 1782) provides insights into its osmoregulation and sex reversal. Genomics. 2021;113:1617–1627. doi: 10.1016/j.ygeno.2021.04.017. - DOI - PubMed
-
- Jia P.Y., Guo H.Y., Zhu K.C., Liu B.S., Guo L., Zhang N., Jiang S.G., Zhang D.C. Cryopreservation of sperm of Acanthopagrus latus. S. China Fish. Sci. 2021;17:58–65. (Abstract in English Only)
LinkOut - more resources
Full Text Sources
Miscellaneous

