Protective Effects of Exogenous Melatonin Administration on White Fat Metabolism Disruption Induced by Aging and a High-Fat Diet in Mice
- PMID: 39765828
- PMCID: PMC11672923
- DOI: 10.3390/antiox13121500
Protective Effects of Exogenous Melatonin Administration on White Fat Metabolism Disruption Induced by Aging and a High-Fat Diet in Mice
Abstract
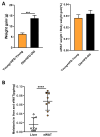
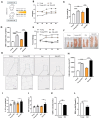
Obesity has emerged as a major risk factor for human health, exacerbated by aging and changes in dietary habits. It represents a significant health challenge, particularly for older people. While numerous studies have examined the effects of obesity and aging on fat metabolism independently, research on their combined effects is limited. In the present study, the protective action against white fat accumulation after a high-fat diet (HFD) exerted by exogenous melatonin, a circadian hormone endowed with antioxidant properties also involved in fat metabolism, was investigated in a mouse model. For this purpose, a battery of tests was applied before and after the dietary and melatonin treatments of the animals, including epididymal white adipose tissue (eWAT) histological evaluations, transcriptomic and lipidomic analyses, real-time PCR tests, immunofluorescence staining, Western blot, the appraisal of serum melatonin levels, and transmission electron microscopy. This study found that aged mice on a high-fat diet (HFD) showed increased lipid deposition, inflammation, and reduced antioxidant glutathione (GSH) levels compared to younger mice. Lipidomic and transcriptomic analyses revealed elevated triglycerides, diglycerides, ceramides, and cholesterol, along with decreased sphingomyelin and fatty acids in eWAT. The genes linked to inflammation, NF-κB signaling, autophagy, and lipid metabolism, particularly the melatonin and glutathione pathways, were significantly altered. The aged HFD mice also exhibited reduced melatonin levels in serum and eWAT. Melatonin supplementation reduced lipid deposition, increased melatonin and GSH levels, and upregulated AANAT and MTNR1A expression in eWAT, suggesting that melatonin alleviates eWAT damage via the MTNR1A pathway. It also suppressed inflammatory markers (e.g., TNF-α, NLRP3, NF-κB, IL-1β, and CEBPB) and preserved mitochondrial function through enhanced mitophagy. This study highlights how aging and HFD affect lipid metabolism and gene expression, offering potential intervention strategies. These findings provide important insights into the mechanisms of fat deposition associated with aging and a high-fat diet, suggesting potential intervention strategies.
Keywords: aging; eWAT; inflammation; lipidomics; melatonin; obesity.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures








References
-
- Benayoun B.A., Pollina E.A., Singh P.P., Mahmoudi S., Harel I., Casey K.M., Dulken B.W., Kundaje A., Brunet A. Remodeling of epigenome and transcriptome landscapes with aging in mice reveals widespread induction of inflammatory responses. Genome Res. 2019;29:697–709. doi: 10.1101/gr.240093.118. - DOI - PMC - PubMed
Grants and funding
- LGD22C040008/This research was funded by grants from the Zhejiang Provincial Natural Science Foundation of China
- 2023GJYY24 and 2023JKJNTZ05/the Research Project of Zhejiang Chinese Medical University
- Y202351395/A Project Supported by Scientific Research Fund of Zhejiang Provincial Education Department
LinkOut - more resources
Full Text Sources

