Identification of Transcriptional Regulators of Immune Evasion Across Cancers: An Alternative Immunotherapeutic Strategy for Cholangiocarcinoma
- PMID: 39766097
- PMCID: PMC11674672
- DOI: 10.3390/cancers16244197
Identification of Transcriptional Regulators of Immune Evasion Across Cancers: An Alternative Immunotherapeutic Strategy for Cholangiocarcinoma
Abstract
Background: Cancer immune evasion is a multifaceted process that synchronizes pro-tumoral immune infiltration, immunosuppressive inflammation, and inhibitory immune checkpoint expression (IC). Current immunotherapies combat this issue by reinstating immunosurveillance of tumors; however, it benefits a limited patient population. Thus, a more effective immunotherapeutic strategy is warranted to cater to specific patient populations. This investigation introduces a novel immunotherapeutic strategy via inhibition of master regulators of immune evasion (MR-IE).
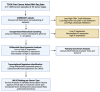
Methods: Samples of the TCGA Pan-Cancer Atlas transcriptomic data were subset and stratified based on IC and estimated immune cell infiltration. Transcriptomic analysis was conducted to unravel pathways associated with the immune evasion process. Transcription factor enrichment and survival analyses were conducted to identify and rank candidate MR-IEs per cancer type.
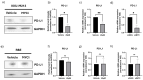
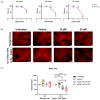
Results: Inhibition of the top-ranking MR-IE candidate of cholangiocarcinoma (CCA), MYC, modulated the gene and protein expression of PD-L1. Moreover, pro-tumoral inflammatory markers, IFNA21 and CX3CL1, were downregulated, and anti-tumoral cytokines, IL-18 and IL-16, were upregulated. Lastly, MYC inhibition potentiated fourth-generation anti-folate receptor alpha (FRα) CAR-T cell therapy against CCA cells.
Conclusions: Cumulatively, this study highlights the promise of MR-IE inhibition as a novel potent immunotherapeutic strategy for the treatment of CCA and offers a candidate list of MR-IEs per cancer type for further validation.
Keywords: cancer; cholangiocarcinoma; immune checkpoints; immunotherapy; transcriptomics.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as potential conflicts of interest.
Figures


References
-
- Gupta I., Hussein O., Sastry K., Bougarn S., Gopinath N., Chin-Smith E., Sinha Y., Korashy H., Maccalli C. Deciphering the complexities of cancer cell immune evasion: Mechanisms and therapeutic implications. Adv. Cancer Biol. Metastasis. 2023;8:100107. doi: 10.1016/j.adcanc.2023.100107. - DOI
-
- Schachter J., Ribas A., Long G.V., Arance A., Grob J.J., Mortier L., Daud A., Carlino M.S., McNeil C., Lotem M., et al. Pembrolizumab versus ipilimumab for advanced melanoma: Final overall survival results of a multicentre, randomised, open-label phase 3 study (KEYNOTE-006) Lancet. 2017;390:1853–1862. doi: 10.1016/S0140-6736(17)31601-X. - DOI - PubMed
Grants and funding
- (FF-062/2566)/Mahidol University (Fundamental Fund: fiscal year 2023 by National Science Research and In-novation Fund (NSRF)
- B36G660002/NSRF via the Program Management Unit for Human Resources and Institutional Development, Research, and Innovation
- 2023/Mahidol International Postdoctoral Fellowship
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

