The Functional Interaction Between Epstein-Barr Virus and MYC in the Pathogenesis of Burkitt Lymphoma
- PMID: 39766110
- PMCID: PMC11674381
- DOI: 10.3390/cancers16244212
The Functional Interaction Between Epstein-Barr Virus and MYC in the Pathogenesis of Burkitt Lymphoma
Abstract
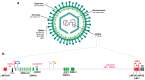
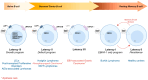
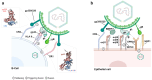
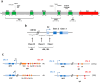
The Epstein-Barr virus (EBV) is associated with a wide range of diseases, malignant and non-malignant. EBV was, in fact, the first virus described with cell transformation capacity, discovered by Epstein in 1964 in lymphoma samples from African children. Since then, EBV has been associated with several human tumors including nasopharyngeal carcinoma, gastric carcinoma, T-cell lymphoma, Hodgkin lymphoma, diffuse large B cell lymphoma, and Burkitt lymphoma among others. The molecular hallmark of Burkitt lymphoma (BL) is a chromosomal translocation that involves the MYC gene and immunoglobulin loci, resulting in the deregulated expression of MYC, an oncogenic transcription factor that appears deregulated in about half of human tumors. The role of MYC in lymphoma is well established, as MYC overexpression drives B cell proliferation through multiple mechanisms, foremost, the stimulation of the cell cycle. Indeed, MYC is found overexpressed or deregulated in several non-Hodgkin lymphomas. Most endemic and many sporadic BLs are associated with EBV infection. While some mechanisms by which EBV can contribute to BL have been reported, the mechanism that links MYC translocation and EBV infection in BL is still under debate. Here, we review the main EBV-associated diseases, with a special focus on BL, and we discuss the interaction of EBV and MYC translocation during B cell malignant transformation in BL.
Keywords: Burkitt lymphoma; Epstein–Barr virus; MYC.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
References
-
- Gratama J.W., Ernberg I. Molecular epidemiology of Epstein-Barr virus infection. Adv. Cancer Res. 1995;67:197–255. - PubMed
-
- de-Thé G., Day N.E., Geser A., Lavoué M.F., Ho J.H., Simons M.J., Sohier R., Tukei P., Vonka V., Zavadova H. Sero-epidemiology of the Epstein-Barr virus: Preliminary analysis of an international study—A review. IARC Sci. Publ. 1975;11:3–16. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

