Integrative Stacking Machine Learning Model for Small Cell Lung Cancer Prediction Using Metabolomics Profiling
- PMID: 39766124
- PMCID: PMC11727543
- DOI: 10.3390/cancers16244225
Integrative Stacking Machine Learning Model for Small Cell Lung Cancer Prediction Using Metabolomics Profiling
Abstract
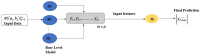
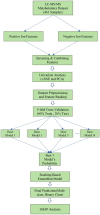
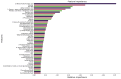
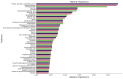
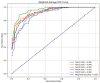
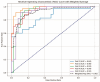
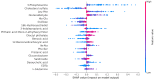
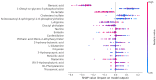
Background: Small cell lung cancer (SCLC) is an extremely aggressive form of lung cancer, characterized by rapid progression and poor survival rates. Despite the importance of early diagnosis, the current diagnostic techniques are invasive and restricted. Methods: This study presents a novel stacking-based ensemble machine learning approach for classifying small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC) using metabolomics data. The analysis included 191 SCLC cases, 173 NSCLC cases, and 97 healthy controls. Feature selection techniques identified significant metabolites, with positive ions proving more relevant. Results: For multi-class classification (control, SCLC, NSCLC), the stacking ensemble achieved 85.03% accuracy and 92.47 AUC using Support Vector Machine (SVM). Binary classification (SCLC vs. NSCLC) further improved performance, with ExtraTreesClassifier reaching 88.19% accuracy and 92.65 AUC. SHapley Additive exPlanations (SHAP) analysis revealed key metabolites like benzoic acid, DL-lactate, and L-arginine as significant predictors. Conclusions: The stacking ensemble approach effectively leverages multiple classifiers to enhance overall predictive performance. The proposed model effectively captures the complementary strengths of different classifiers, enhancing the detection of SCLC and NSCLC. This work accentuates the potential of combining metabolomics with advanced machine learning for non-invasive early lung cancer subtype detection, offering an alternative to conventional biopsy methods.
Keywords: NSCLC; SCLC; machine learning; serum metabolomics; stacking ensemble model.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources

