A Proteomic Examination of Plasma Extracellular Vesicles Across Colorectal Cancer Stages Uncovers Biological Insights That Potentially Improve Prognosis
- PMID: 39766158
- PMCID: PMC11674649
- DOI: 10.3390/cancers16244259
A Proteomic Examination of Plasma Extracellular Vesicles Across Colorectal Cancer Stages Uncovers Biological Insights That Potentially Improve Prognosis
Abstract
Background: Recent advancements in understanding plasma extracellular vesicles (EVs) and their role in disease biology have provided additional unique insights into the study of Colorectal Cancer (CRC).
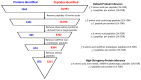
Methods: This study aimed to gain biological insights into disease progression from plasma-derived extracellular vesicle proteomic profiles of 80 patients (20 from each CRC stage I-IV) against 20 healthy age- and sex-matched controls using a high-resolution SWATH-MS proteomics with a reproducible centrifugation method to isolate plasma EVs.
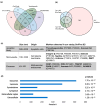
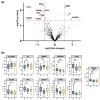
Results: We applied the High-Stringency Human Proteome Project (HPP) guidelines for SWATH-MS analysis, which refined our initial EV protein identification from 1362 proteins (10,993 peptides) to a more reliable and confident subset of 853 proteins (6231 peptides). In early-stage CRC, we identified 11 plasma EV proteins with differential expression between patients and healthy controls (three up-regulated and eight down-regulated), many of which are involved in key cancer hallmarks. Additionally, within the same cohort, we analysed EV proteins associated with tumour recurrence to identify potential prognostic indicators for CRC. A subset of up-regulated proteins associated with extracellular vesicle formation (GDI1, NSF, and TMED9) and the down-regulation of TSG101 suggest that micro-metastasis may have occurred earlier than previously anticipated.
Discussion: By employing stringent proteomic analysis and a robust SWATH-MS approach, we identified dysregulated EV proteins that potentially indicate early-stage CRC and predict recurrence risk, including proteins involved in metabolism, cytoskeletal remodelling, and immune response. While our findings underline discrepancies with other studies due to differing isolation and stringency parameters, they provide valuable insights into the complexity of the EV proteome, emphasising the need for standardised protocols and larger, well-controlled studies to validate potential biomarkers.
Keywords: SWATH-MS; colorectal cancer; exosomes; extracellular vesicles; plasma microvesicle proteins; protein biomarkers.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- Araghi M., Soerjomataram I., Bardot A., Ferlay J., Cabasag C.J., Morrison D.S., De P., Tervonen H., Walsh P.M., Bucher O., et al. Changes in colorectal cancer incidence in seven high-income countries: A population-based study. Lancet Gastroenterol. Hepatol. 2019;4:511–518. doi: 10.1016/S2468-1253(19)30147-5. - DOI - PMC - PubMed
-
- Alnakli A.A.A., Mohamedali A., Heng B., Chan C., Shin J.-S., Solomon M., Chapuis P., Guillemin G.J., Baker M.S., Ahn S.B. Protein prognostic biomarkers in stage II colorectal cancer: Implications for post-operative management. BJC Rep. 2024;2:13. doi: 10.1038/s44276-024-00043-z. - DOI - PMC - PubMed
-
- Young P.E., Womeldorph C.M., Johnson E.K., Maykel J.A., Brucher B., Stojadinovic A., Avital I., Nissan A., Steele S.R. Early detection of colorectal cancer recurrence in patients undergoing surgery with curative intent: Current status and challenges. J. Cancer. 2014;5:262–271. doi: 10.7150/jca.7988. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

