Disrupted Lipid Metabolism, Cytokine Signaling, and Dormancy: Hallmarks of Doxorubicin-Resistant Triple-Negative Breast Cancer Models
- PMID: 39766172
- PMCID: PMC11674486
- DOI: 10.3390/cancers16244273
Disrupted Lipid Metabolism, Cytokine Signaling, and Dormancy: Hallmarks of Doxorubicin-Resistant Triple-Negative Breast Cancer Models
Abstract
Background: Chemoresistance in triple-negative breast cancer (TNBC) presents a significant clinical hurdle, limiting the efficacy of treatments like doxorubicin. This study aimed to explore the molecular changes associated with doxorubicin resistance and identify potential therapeutic targets to overcome this resistance, thereby improving treatment outcomes for TNBC patients.
Methods: Doxorubicin-resistant (DoxR) TNBC models (MDA-MB-231 and BT-549) were generated by exposing cells to increasing concentrations of doxorubicin. RNA sequencing (RNA-Seq) was performed using the Illumina platform, followed by bioinformatics analysis with CLC Genomics Workbench and iDEP. Functional assays assessed proliferation, sphere formation, migration, and cell cycle changes. Protein expression and phosphorylation were confirmed via Western blotting. Pathway and network analyses were conducted using Ingenuity Pathway Analysis (IPA) and STRING, while survival analysis was performed using Kaplan-Meier Plotter database.
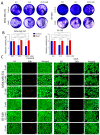
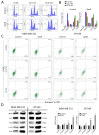
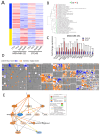
Results: DoxR cells exhibited reduced proliferation, sphere formation, and migration, but showed enhanced tolerance to doxorubicin. Increased CHK2 and p53 phosphorylation indicated cellular dormancy as a resistance mechanism. RNA-Seq analysis revealed upregulation of cytokine signaling and stress-response pathways, while cholesterol and lipid biosynthesis were suppressed. Activation of the IL1β cytokine network was prominent in DoxR cells, and CRISPR-Cas9 screens data identified dependencies on genes involved in rRNA biogenesis and metabolism. A 27-gene signature associated with doxorubicin resistance was linked to worse clinical outcomes in a large breast cancer cohort (HR = 1.76, FDR p < 2.0 × 10-13).
Conclusions: This study uncovers potential therapeutic strategies for overcoming TNBC resistance, including dormancy reversal and targeting onco-ribosomal pathways and cytokine signaling networks, to improve the efficacy of doxorubicin-based treatments.
Keywords: cellular dormancy; chemoresistance; doxorubicin; therapeutic strategies; triple-negative breast cancer (TNBC).
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Garrido-Castro A.C., Lin N.U., Polyak K. Insights into Molecular Classifications of Triple-Negative Breast Cancer: Improving Patient Selection for Treatment. Cancer Discov. 2019;9:176–198. doi: 10.1158/2159-8290.CD-18-1177. - DOI - PMC - PubMed
-
- Santonja A., Sanchez-Munoz A., Lluch A., Chica-Parrado M.R., Albanell J., Chacon J.I., Antolin S., Jerez J.M., de la Haba J., de Luque V., et al. Triple negative breast cancer subtypes and pathologic complete response rate to neoadjuvant chemotherapy. Oncotarget. 2018;9:26406–26416. doi: 10.18632/oncotarget.25413. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

