Thyroid Carcinoma Glycoproteins Express Altered N-Glycans with 3-O-Sulfated Galactose Residues
- PMID: 39766189
- PMCID: PMC11727208
- DOI: 10.3390/biom14121482
Thyroid Carcinoma Glycoproteins Express Altered N-Glycans with 3-O-Sulfated Galactose Residues
Abstract
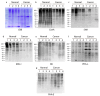
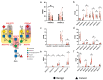
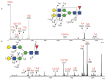
Aberrant protein glycosylation is a hallmark alteration of cancer and is highly associated with cancer progression. Papillary thyroid cancer (PTC) is the most common type of thyroid cancer, but the N-glycosylation of its glycoproteins has not been well characterized. In this work, we analyzed multiple freshly prepared PTC specimens along with paired normal tissue obtained from thyroidectomies. Glycomic analyses focused on Asn-linked (N)-glycans and employed mass spectrometry (MS), along with Western blot approaches of total solubilized materials that were examined for binding by specific lectins and a monoclonal antibody (mAb) O6, specific for 3-O-sulfated galactose residues. We observed major differences in PTC versus paired normal specimens, as PTC specimens exhibited higher levels of N-glycan branching and bisection with N-acetylglucosamine residues, consistent with RNAseq data. We also found that 3-O-sulfated galactose was present in N-glycans of multiple glycoproteins from both PTC and control specimens, as recognized by the O6 mAb and as confirmed by MS analyses. These results provide new insights into the N-glycans present in glycoproteins of thyroid cancer and context for further studies of these altered glycans as biomarkers and targets for therapeutics.
Keywords: GlcNAc bisection; RNAseq; glycosylation; lectin histochemistry; mass spectrometry; papillary thyroid cancer; sialylation; sulfation; sulfoglycomics; thyroid carcinoma; western blotting.
Conflict of interest statement
The authors declare that there are no conflicts of interest among any of the authors.
Figures



Similar articles
-
Novel lamprey antibody recognizes terminal sulfated galactose epitopes on mammalian glycoproteins.Commun Biol. 2021 Jun 3;4(1):674. doi: 10.1038/s42003-021-02199-7. Commun Biol. 2021. PMID: 34083726 Free PMC article.
-
N-glycan profiling of papillary thyroid carcinoma tissues by MALDI-TOF-MS.Anal Biochem. 2019 Nov 1;584:113389. doi: 10.1016/j.ab.2019.113389. Epub 2019 Aug 7. Anal Biochem. 2019. PMID: 31400301
-
Quantitative O-glycomics based on improvement of the one-pot method for nonreductive O-glycan release and simultaneous stable isotope labeling with 1-(d0/d5)phenyl-3-methyl-5-pyrazolone followed by mass spectrometric analysis.J Proteomics. 2017 Jan 6;150:18-30. doi: 10.1016/j.jprot.2016.08.012. Epub 2016 Aug 29. J Proteomics. 2017. PMID: 27585995
-
The glycosylation landscape of prostate cancer tissues and biofluids.Adv Cancer Res. 2024;161:1-30. doi: 10.1016/bs.acr.2024.04.005. Epub 2024 Apr 25. Adv Cancer Res. 2024. PMID: 39032948 Review.
-
Analysis of N- and O-linked site-specific glycosylation by ion mobility mass spectrometry: State of the art and future directions.Proteomics. 2024 Jun;24(12-13):e2300281. doi: 10.1002/pmic.202300281. Epub 2024 Jan 3. Proteomics. 2024. PMID: 38171879 Review.
Cited by
-
Glycomics and Glycoproteomics Reveal Distinct Oligomannose Carriers Across Bladder Cancer Stages.Int J Mol Sci. 2025 May 20;26(10):4891. doi: 10.3390/ijms26104891. Int J Mol Sci. 2025. PMID: 40430030 Free PMC article.
References
-
- Bellis S.L., Reis C.A., Varki A., Kannagi R., Stanley P. Glycosylation Changes in Cancer. In: Varki A., Cummings R.D., Esko J.D., Stanley P., Hart G.W., Aebi M., Mohnen D., Kinoshita T., Packer N.H., Prestegard J.H., et al., editors. Essentials of Glycobiology. 4th ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY, USA: 2022. pp. 631–644.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

