NALCN Promoter Methylation as a Biomarker for Metastatic Risk in a Cohort of Non-Small Cell Lung Cancer Patients
- PMID: 39766220
- PMCID: PMC11673096
- DOI: 10.3390/biom14121514
NALCN Promoter Methylation as a Biomarker for Metastatic Risk in a Cohort of Non-Small Cell Lung Cancer Patients
Abstract
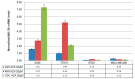
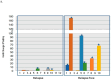
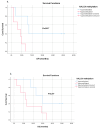
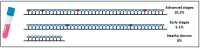
Liquid biopsy enables real-time monitoring of tumor development and response to therapy through the analysis of CTCs and ctDNA. NALCN is a sodium leak channel that is frequently involved in tumor evolution and immunity and acts as a tumor suppressor. Deletion of NALCN has been shown to increase cancer metastasis and the number of CTCs in peripheral blood. In this study, we investigated for the first time NALCN promoter methylation in (a) Aza-treated cell lines (A549, TE671, BT20, and MDA-MB-468), (b) paired NSCLC tissues (n = 22), and (c) plasma cell-free DNA (ctDNA) from patients with NSCLC (early stage n = 39, metastatic n = 39) and DNA from 10 healthy donors (HD) using a newly developed highly specific and sensitive real-time MSP method. Treatment with 5'-aza-dC induced the expression of NALCN only in the A549 cell line, suggesting that DNA methylation regulates its expression in certain cancers. The mRNA expression levels of NALCN were quantified in non-small cell lung cancer (NSCLC) and adjacent non-cancerous tissues, and it was found to be underexpressed in 54.5% of tumor tissues, with significantly higher expression in recurrence-free patients (p = 0.009) than in patients who relapsed. The NALCN methylation level was not statisticallysignificantlycorrelated with the corresponding expression (p = 0.439), while Kaplan-Meier analysis showed an association between NALCN promoter hypermethylation and worse disease-free intervals (DFIs) (p = 0.017). Evaluation of NALCN methylation in ctDNA revealed that it was detected in 5.1% of early and 10.2% of advanced cases. Our results strongly suggest that epigenetic inactivation of NALCN may be a predictor of metastasis in NSCLC. Our results should be validated in further studies based on a larger patient cohort to further investigate whether DNA methylation of the NALCN promoter could serve as a potential prognostic DNA methylation biomarker and predictor of metastasis in NSCLC.
Keywords: NALCN; NSCLC; metastasis; methylation; prognostic biomarker.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

