Affinity Tag-Free Purification of SARS-CoV-2 N Protein and Its Crystal Structure in Complex with ssDNA
- PMID: 39766245
- PMCID: PMC11673995
- DOI: 10.3390/biom14121538
Affinity Tag-Free Purification of SARS-CoV-2 N Protein and Its Crystal Structure in Complex with ssDNA
Abstract
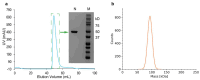
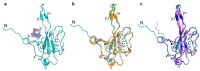
The nucleocapsid (N) protein is one of the four structural proteins in SARS-CoV-2, playing key roles in viral assembly, immune evasion, and stability. One of its primary functions is to protect viral RNA by forming the nucleocapsid. However, the precise mechanisms by which the N protein interacts with viral RNA and assembles into a nucleocapsid remain unclear. Compared to other SARS-CoV-2 components, targeting the N protein has several advantages: it exhibits higher sequence conservation, lower mutation rates, and stronger immunogenicity, making it an attractive target for antiviral drug development and diagnostics. Therefore, a detailed understanding of the N protein's structure is essential for deciphering its role in viral assembly and developing effective therapeutics. In this study, we report the expression and purification of a soluble recombinant N protein, along with a 1.55 Å resolution crystal structure of its nucleic acid-binding domain (N-NTD) in complex with ssDNA. Our structure revealed new insights into the conformation and interaction of the flexible N-arm, which could aid in understanding nucleocapsid assembly. Additionally, we identified residues that are critical for ssDNA interaction.
Keywords: COVID-19; N-terminal domain; SARS-CoV-2; X-ray crystallography; atomic-resolution structure; mass photometry; nucleocapsid protein; protein expression; protein purification; protein–DNA interactions.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
-
- Aleem A., Akbar Samad A.B., Vaqar S. StatPearls. StatPearls Publishing; Treasure Island, FL, USA: 2024. Emerging Variants of SARS-CoV-2 and Novel Therapeutics Against Coronavirus (COVID-19) - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

