Proteomics: An Essential Tool to Study Plant-Specialized Metabolism
- PMID: 39766246
- PMCID: PMC11674799
- DOI: 10.3390/biom14121539
Proteomics: An Essential Tool to Study Plant-Specialized Metabolism
Abstract
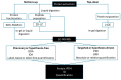
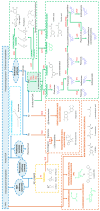
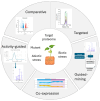
Plants are a valuable source of specialized metabolites that provide a plethora of therapeutic applications. They are natural defenses that plants use to adapt and respond to their changing environment. Decoding their biosynthetic pathways and understanding how specialized plant metabolites (SPMs) respond to biotic or abiotic stress will provide vital knowledge for plant biology research and its application for the future sustainable production of many SPMs of interest. Here, we focus on the proteomic approaches and strategies that help with the study of plant-specialized metabolism, including the: (i) discovery of key enzymes and the clarification of their biosynthetic pathways; (ii) study of the interconnection of both primary (providers of carbon and energy for SPM production) and specialized (secondary) metabolism; (iii) study of plant responses to biotic and abiotic stress; (iv) study of the regulatory mechanisms that direct their biosynthetic pathways. Proteomics, as exemplified in this review by the many studies performed to date, is a powerful tool that forms part of omics-driven research. The proteomes analysis provides an additional unique level of information, which is absent from any other omics studies. Thus, an integrative analysis, considered versus a single omics analysis, moves us more closely toward a closer interpretation of real cellular processes. Finally, this work highlights advanced proteomic technologies with immediate applications in the field.
Keywords: biosynthesis; mass spectrometry; natural products or compounds; proteomics; secondary metabolism; specialized plant metabolite.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
References
-
- Stander E.A., Lehka B., Carqueijeiro I., Cuello C., Hansson F.G., Jansen H.J., Dugé De Bernonville T., Birer Williams C., Vergès V., Lezin E., et al. The Rauvolfia Tetraphylla Genome Suggests Multiple Distinct Biosynthetic Routes for Yohimbane Monoterpene Indole Alkaloids. Commun. Biol. 2023;6:1197. doi: 10.1038/s42003-023-05574-8. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

