Core-Shell PLGA Nanoparticles: In Vitro Evaluation of System Integrity
- PMID: 39766308
- PMCID: PMC11674307
- DOI: 10.3390/biom14121601
Core-Shell PLGA Nanoparticles: In Vitro Evaluation of System Integrity
Abstract
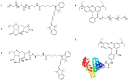
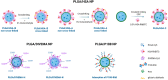
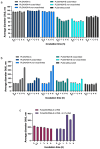
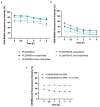
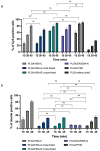
The objective of this study was to compare the properties of core-shell nanoparticles with a PLGA core and shells composed of different types of polymers, focusing on their structural integrity. The core PLGA nanoparticles were prepared either through a high-pressure homogenization-solvent evaporation technique or nanoprecipitation, using poloxamer 188 (P188), a copolymer of divinyl ether with maleic anhydride (DIVEMA), and human serum albumin (HSA) as the shell-forming polymers. The shells were formed through adsorption, interfacial embedding, or conjugation. For dual fluorescent labeling, the core- and shell-forming polymers were conjugated with Cyanine5, Cyanine3, and rhodamine B. The nanoparticles had negative zeta potentials and sizes ranging from 100 to 250 nm (measured using DLS) depending on the shell structure and preparation technique. The core-shell structure was confirmed using TEM and fluorescence spectroscopy, with the appearance of FRET phenomena due to the donor-acceptor properties of the labels. All of the shells enhanced the cellular uptake of the nanoparticles in Gl261 murine glioma cells. The integrity of the core-shell structures upon their incubation with the cells was evidenced by intracellular colocalization of the fluorescent labels according to the Manders' colocalization coefficients. This comprehensive approach may be useful for the selection of the optimal preparation method even at the early stages of the core-shell nanoparticle development.
Keywords: DIVEMA; Gl261 cells; HSA; PLGA; confocal microscopy; core–shell nanoparticles; poloxamer 188.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



Similar articles
-
Release kinetics of fluorescent dyes from PLGA nanoparticles in retinal blood vessels: In vivo monitoring and ex vivo localization.Eur J Pharm Biopharm. 2020 May;150:131-142. doi: 10.1016/j.ejpb.2020.03.006. Epub 2020 Mar 6. Eur J Pharm Biopharm. 2020. PMID: 32151727
-
Lipid-PLGA hybrid nanoparticles of paclitaxel: Preparation, characterization, in vitro and in vivo evaluation.Mater Sci Eng C Mater Biol Appl. 2020 Apr;109:110576. doi: 10.1016/j.msec.2019.110576. Epub 2019 Dec 21. Mater Sci Eng C Mater Biol Appl. 2020. PMID: 32228957
-
BSA-PLGA-based core-shell nanoparticles as carrier system for water-soluble drugs.Pharm Res. 2013 Sep;30(9):2396-409. doi: 10.1007/s11095-013-1084-6. Epub 2013 Jun 12. Pharm Res. 2013. PMID: 23756758
-
The effect of poloxamer 188 on nanoparticle morphology, size, cancer cell uptake, and cytotoxicity.Nanomedicine. 2010 Feb;6(1):170-8. doi: 10.1016/j.nano.2009.05.004. Epub 2009 May 15. Nanomedicine. 2010. PMID: 19447200
-
Comparative whole corona fingerprinting and protein adsorption thermodynamics of PLGA and PCL nanoparticles in human serum.Colloids Surf B Biointerfaces. 2020 Apr;188:110816. doi: 10.1016/j.colsurfb.2020.110816. Epub 2020 Jan 22. Colloids Surf B Biointerfaces. 2020. PMID: 31991290 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

