Dissecting Cytophagalysin: Structural and Biochemical Studies of a Bacterial Pappalysin-Family Metallopeptidase
- PMID: 39766312
- PMCID: PMC11674741
- DOI: 10.3390/biom14121604
Dissecting Cytophagalysin: Structural and Biochemical Studies of a Bacterial Pappalysin-Family Metallopeptidase
Abstract
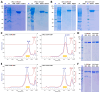
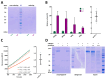
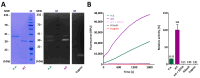
Cytophaga is a genus of Gram-negative bacteria occurring in soil and the gut microbiome. It is closely related to pathogenic Flavobacterium spp. that cause severe diseases in fish. Cytophaga strain L43-1 secretes cytophagalysin (CPL1), a 137 kDa peptidase with reported collagenolytic and gelatinolytic activity. We performed highly-confident structure prediction calculations for CPL1, which identified 11 segments and domains, including a signal peptide for secretion, a prosegment (PS) for latency, a metallopeptidase (MP)-like catalytic domain (CD), and eight immunoglobulin (Ig)-like domains (D3-D10). In addition, two short linkers were found at the D8-D9 and D9-D10 junctions, and the structure would be crosslinked by four disulfide bonds. The CPL1 CD was found closest to ulilysin from Methanosarcina acetivorans, which assigns CPL1 to the lower-pappalysin family within the metzincin clan of MPs. Based on the structure predictions, we aimed to produce constructs spanning the full-length enzyme, as well as PS+CD, PS+CD+D3, and PS+CD+D3+D4. However, we were successful only with the latter three constructs. We could activate recombinant CPL1 by PS removal employing trypsin, and found that both zymogen and mature CPL1 were active in gelatin zymography and against a fluorogenic gelatin variant. This activity was ablated in a mutant, in which the catalytic glutamate described for lower pappalyins and other metzincins was replaced by alanine, and by a broad-spectrum metal chelator. Overall, these results proved that our recombinant CPL1 is a functional active MP, thus supporting the conclusions derived from the structure predictions.
Keywords: functional characterization; metallopeptidase; pappalysin family; protease activation; recombinant protein expression.
Conflict of interest statement
The authors declare no financial or non-financial conflicts of interest with the contents of this article, and the funders had no involvement in the study’s design, data collection, analysis, or interpretation, manuscript preparation, or the decision to publish the findings.
Figures



Similar articles
-
Structural insights into latency of the metallopeptidase ulilysin (lysargiNase) and its unexpected inhibition by a sulfonyl-fluoride inhibitor of serine peptidases.Dalton Trans. 2023 Mar 21;52(12):3610-3622. doi: 10.1039/d3dt00458a. Dalton Trans. 2023. PMID: 36857690
-
Substrate specificity of a metalloprotease of the pappalysin family revealed by an inhibitor and a product complex.Arch Biochem Biophys. 2007 Jan 1;457(1):57-72. doi: 10.1016/j.abb.2006.10.004. Epub 2006 Oct 24. Arch Biochem Biophys. 2007. PMID: 17097044
-
Molecular analysis of ulilysin, the structural prototype of a new family of metzincin metalloproteases.J Biol Chem. 2006 Jun 30;281(26):17920-8. doi: 10.1074/jbc.M600907200. Epub 2006 Apr 20. J Biol Chem. 2006. PMID: 16627477
-
Architecture and function of metallopeptidase catalytic domains.Protein Sci. 2014 Feb;23(2):123-44. doi: 10.1002/pro.2400. Protein Sci. 2014. PMID: 24596965 Free PMC article. Review.
-
Multiple Architectures and Mechanisms of Latency in Metallopeptidase Zymogens.Chem Rev. 2018 Jun 13;118(11):5581-5597. doi: 10.1021/acs.chemrev.8b00030. Epub 2018 May 18. Chem Rev. 2018. PMID: 29775286 Review.
References
-
- Fushimi N., Ee C.E., Nakajima T., Ichishima E. Aspzincin, a Family of Metalloendopeptidases with a New Zinc-Binding Motif. Identification of New Zinc-Binding Sites (His128, His132, and Asp164) and Three Catalytically Crucial Residues (Glu129, Asp143, and Tyr106) of Deuterolysin from Aspergillus Oryzae by Site-Directed Mutagenesis. J. Biol. Chem. 1999;274:24195–24201. doi: 10.1074/jbc.274.34.24195. - DOI - PubMed
-
- Bode W., Gomis-Rüth F.X., Stöcker W. Astacins, Serralysins, Snake Venom and Matrix Metalloproteinases Exhibit Identical Zinc-Binding Environments (HEXXHXXGXXH and Met-Turn) and Topologies and Should Be Grouped into a Common Family, the ‘metzincins’. FEBS Lett. 1993;331:134–140. doi: 10.1016/0014-5793(93)80312-I. - DOI - PubMed
MeSH terms
Substances
Grants and funding
- RYC2020-029773-I/Ministerio de Ciencia, Innovación y Universidades
- PID2021-128682OA-I00/Ministerio de Ciencia, Innovación y Universidades
- PRE2020-096731/Ministerio de Ciencia, Innovación y Universidades
- JAEINT_22_02654/Consejo Superior de Investigaciones Científicas
- 2021SGR00423/Government of Catalonia
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

