Molecular Pathways Linking High-Fat Diet and PM2.5 Exposure to Metabolically Abnormal Obesity: A Systematic Review and Meta-Analysis
- PMID: 39766314
- PMCID: PMC11674716
- DOI: 10.3390/biom14121607
Molecular Pathways Linking High-Fat Diet and PM2.5 Exposure to Metabolically Abnormal Obesity: A Systematic Review and Meta-Analysis
Abstract
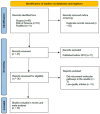
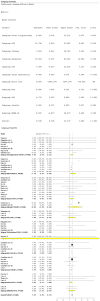
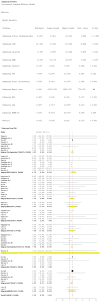
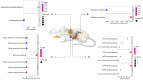
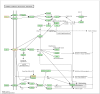
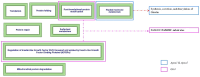
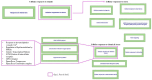
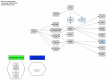
Obesity, influenced by environmental pollutants, can lead to complex metabolic disruptions. This systematic review and meta-analysis examined the molecular mechanisms underlying metabolically abnormal obesity caused by exposure to a high-fat diet (HFD) and fine particulate matter (PM2.5). Following the PRISMA guidelines, articles from 2019 to 2024 were gathered from Scopus, Web of Science, and PubMed, and a random-effects meta-analysis was performed, along with subgroup analyses and pathway enrichment analyses. This study was registered in the Open Science Framework. Thirty-three articles, mainly case-control studies and murine models, were reviewed, and they revealed that combined exposure to HFD and PM2.5 resulted in the greatest weight gain (82.835 g, p = 0.048), alongside increases in high-density lipoproteins, insulin, and the superoxide dismutase. HFD enriched pathways linked to adipocytokine signaling in brown adipose tissue, while PM2.5 impacted genes associated with fat formation. Both exposures downregulated protein metabolism pathways in white adipose tissue and activated stress-response pathways in cardiac tissue. Peroxisome proliferator-activated receptor and AMP-activated protein kinase signaling pathways in the liver were enriched, influencing non-alcoholic fatty liver disease. These findings highlight that combined exposure to HFD and PM2.5 amplifies body weight gain, oxidative stress, and metabolic dysfunction, suggesting a synergistic interaction with significant implications for metabolic health.
Keywords: airborne particulate matter; gene–environment interaction; high-fat diet; metabolic pathways; obesity; oxidative stress; signaling pathways.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



Similar articles
-
Effects of a maternal high-fat diet on adipose tissue in murine offspring: A systematic review and meta-analysis.Biochimie. 2022 Oct;201:18-32. doi: 10.1016/j.biochi.2022.06.009. Epub 2022 Jun 30. Biochimie. 2022. PMID: 35779649
-
Combined exercise training and dietary interventions versus independent effect of exercise on ectopic fat in individuals with overweight and obesity: a systematic review, meta-analysis, and meta-regression.J Int Soc Sports Nutr. 2025 Dec;22(1):2528534. doi: 10.1080/15502783.2025.2528534. Epub 2025 Jul 6. J Int Soc Sports Nutr. 2025. PMID: 40619882 Free PMC article.
-
Dietary n-6:n-3 PUFA Ratio Modulates Inflammation-Related Gene Expression and Influences Improvements in Biochemical Parameters in a Murine Model of Diet-Induced Obesity.Nutrients. 2025 Jun 13;17(12):1996. doi: 10.3390/nu17121996. Nutrients. 2025. PMID: 40573106 Free PMC article.
-
Ablation of FAM210A in Brown Adipocytes of Mice Exacerbates High-Fat Diet-Induced Metabolic Dysfunction.Diabetes. 2025 Mar 1;74(3):282-294. doi: 10.2337/db24-0294. Diabetes. 2025. PMID: 39602358
-
The role of gut microbiota in Tirzepatide-mediated alleviation of high-fat diet-induced obesity.Eur J Pharmacol. 2025 Sep 5;1002:177827. doi: 10.1016/j.ejphar.2025.177827. Epub 2025 Jun 12. Eur J Pharmacol. 2025. PMID: 40516844
References
-
- Löffler M.C., Betz M.J., Blondin D.P., Augustin R., Sharma A.K., Tseng Y.-H., Scheele C., Zimdahl H., Mark M., Hennige A.M., et al. Challenges in tackling energy expenditure as obesity therapy: From preclinical models to clinical application. Mol. Metab. 2021;51:101237. doi: 10.1016/j.molmet.2021.101237. - DOI - PMC - PubMed
-
- Mayoral L.P.-C., Andrade G.M., Mayoral E.P.-C., Huerta T.H., Canseco S.P., Rodal Canales F.J., Cabrera-Fuentes H.A., Cruz M.M., Pérez Santiago A.D., Alpuche J.J., et al. Obesity subtypes, related biomarkers & heterogeneity. Indian J. Med. Res. 2020;151:11–21. doi: 10.4103/ijmr.IJMR_1768_17. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

