Metatranscriptomic Analysis Reveals Actively Expressed Antimicrobial-Resistant Genes and Their Hosts in Hospital Wastewater
- PMID: 39766512
- PMCID: PMC11672649
- DOI: 10.3390/antibiotics13121122
Metatranscriptomic Analysis Reveals Actively Expressed Antimicrobial-Resistant Genes and Their Hosts in Hospital Wastewater
Abstract
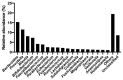
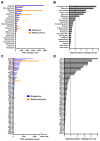
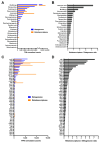
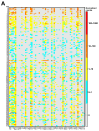
Antimicrobial resistance is a major global concern and economic threat, necessitating a reliable monitoring approach to understand its frequency and spread via the environment. Hospital wastewater serves as a critical reservoir for antimicrobial-resistant organisms; however, its role in resistance gene distribution and dissemination remains poorly understood. This study integrates metagenomic and metatranscriptomic analyses, elucidating the dynamics of antimicrobial resistance in hospital wastewater. Integrated metagenomic and metatranscriptomic sequencing were used to identify actively expressed antimicrobial-resistant genes and antimicrobial-resistant bacteria, offering comprehensive insights into antimicrobial resistance dynamics in hospital wastewater. Liquid chromatography-tandem mass spectrometry analysis revealed the presence of ampicillin, sulbactam, levofloxacin, sulfamethoxazole, and trimethoprim in the sample, which could apply selective pressure on antimicrobial resistance gene expression. While multidrug resistance genes were the most prevalent sequences in both metagenome-assembled genomes and plasmids, plasmid-derived sequences showed a high mRNA/DNA ratio, emphasizing the presence of functionally expressed antimicrobial resistance genes on plasmids rather than on chromosomes. The metagenomic and metatranscriptomic analyses revealed Serratia nevei MAG14 with high mRNA levels of antimicrobial resistance genes; moreover, multidrug-resistant Serratia sp., genetically related to MAG14, was isolated from the wastewater, supporting the phenotypic characterization of crucial antimicrobial-resistant bacteria and validating the genome analysis results. The findings underscore key genes and bacteria as targets for antimicrobial resistance surveillance in hospital wastewater to protect public and environmental health.
Keywords: antimicrobial resistance; hospital wastewater; metatranscriptomic analysis.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study, in the collection, analysis, or interpretation of data, in the writing of the manuscript, or in the decision to publish the results.
Figures



Similar articles
-
Whole genome analysis of multidrug-resistant Escherichia coli isolate collected from drinking water in Armenia revealed the plasmid-borne mcr-1.1-mediated colistin resistance.Microbiol Spectr. 2024 Oct 3;12(10):e0075124. doi: 10.1128/spectrum.00751-24. Epub 2024 Aug 21. Microbiol Spectr. 2024. PMID: 39166856 Free PMC article.
-
Genome-centric analyses of 165 metagenomes show that mobile genetic elements are crucial for the transmission of antimicrobial resistance genes to pathogens in activated sludge and wastewater.Microbiol Spectr. 2024 Mar 5;12(3):e0291823. doi: 10.1128/spectrum.02918-23. Epub 2024 Jan 30. Microbiol Spectr. 2024. PMID: 38289113 Free PMC article.
-
Genomic Analysis of Hospital Plumbing Reveals Diverse Reservoir of Bacterial Plasmids Conferring Carbapenem Resistance.mBio. 2018 Feb 6;9(1):e02011-17. doi: 10.1128/mBio.02011-17. mBio. 2018. PMID: 29437920 Free PMC article.
-
Environmental antimicrobial resistance and its drivers: a potential threat to public health.J Glob Antimicrob Resist. 2021 Dec;27:101-111. doi: 10.1016/j.jgar.2021.08.001. Epub 2021 Aug 25. J Glob Antimicrob Resist. 2021. PMID: 34454098 Review.
-
Antimicrobial Resistance Profile by Metagenomic and Metatranscriptomic Approach in Clinical Practice: Opportunity and Challenge.Antibiotics (Basel). 2022 May 13;11(5):654. doi: 10.3390/antibiotics11050654. Antibiotics (Basel). 2022. PMID: 35625299 Free PMC article. Review.
References
-
- Hanna N., Tamhankar A.J., Stålsby Lundborg C. Antibiotic concentrations and antibiotic resistance in aquatic environments of the WHO Western Pacific and South-East Asia regions: A systematic review and probabilistic environmental hazard assessment. Lancet Planet Health. 2023;7:e45–e54. doi: 10.1016/S2542-5196(22)00254-6. - DOI - PubMed
-
- Milligan E.G., Calarco J., Davis B.C., Keenum I.M., Liguori K., Pruden A., Harwood V.J. A systematic review of culture-based methods for monitoring antibiotic-resistant Acinetobacter, Aeromonas, and Pseudomonas as environmentally relevant pathogens in wastewater and surface water. Curr. Environ. Health Rep. 2023;10:154–171. doi: 10.1007/s40572-023-00393-9. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

