Current Knowledge on CRISPR Strategies Against Antimicrobial-Resistant Bacteria
- PMID: 39766530
- PMCID: PMC11672446
- DOI: 10.3390/antibiotics13121141
Current Knowledge on CRISPR Strategies Against Antimicrobial-Resistant Bacteria
Abstract
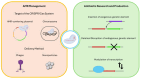
CRISPR/Cas systems have emerged as valuable tools to approach the problem of antimicrobial resistance by either sensitizing or lysing resistant bacteria or by aiding in antibiotic development, with successful applications across diverse organisms, including bacteria and fungi. CRISPR/Cas systems can target plasmids or the bacterial chromosome of AMR-bacteria, and it is especially necessary to have an efficient entry into the target cells, which can be achieved through nanoparticles or bacteriophages. Regarding antibiotic development and production, though the use of CRISPR/Cas in this field is still modest, there is an untapped reservoir of bacterial and fungal natural products, with over 95% yet to be characterized. In Streptomyces, a key antibiotic-producing bacterial genus, CRISPR/Cas has been successfully used to activate silent biosynthetic gene clusters, leading to the discovery of new antibiotics. CRISPR/Cas is also applicable to non-model bacteria and different species of fungi, making it a versatile tool for natural products discovery. Moreover, CRISPR/Cas-based studies offer insights into metabolic regulation and biosynthetic pathways in both bacteria and fungi, highlighting its utility in understanding genetic regulation and improving industrial strains. In this work, we review ongoing innovations on ways to treat antimicrobial resistances and on antibiotic discovery using CRISPR/Cas platforms, highlighting the role of bacteria and fungi in these processes.
Keywords: AMR; BGCs; CRISPR/Cas; Streptomyces; antibiotic; multidrug-resistant bacteria; nanoparticles; phages.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
Advancements in CRISPR-Cas-based strategies for combating antimicrobial resistance.Microbiol Res. 2025 Sep;298:128232. doi: 10.1016/j.micres.2025.128232. Epub 2025 May 22. Microbiol Res. 2025. PMID: 40440869 Review.
-
CRISPR-Cas-Based Antimicrobials: Design, Challenges, and Bacterial Mechanisms of Resistance.ACS Infect Dis. 2023 Jul 14;9(7):1283-1302. doi: 10.1021/acsinfecdis.2c00649. Epub 2023 Jun 22. ACS Infect Dis. 2023. PMID: 37347230 Free PMC article. Review.
-
Activating natural product synthesis using CRISPR interference and activation systems in Streptomyces.Nucleic Acids Res. 2022 Jul 22;50(13):7751-7760. doi: 10.1093/nar/gkac556. Nucleic Acids Res. 2022. PMID: 35801861 Free PMC article.
-
CRISPR-Cas System: A New Dawn to Combat Antibiotic Resistance.BioDrugs. 2024 May;38(3):387-404. doi: 10.1007/s40259-024-00656-3. Epub 2024 Apr 11. BioDrugs. 2024. PMID: 38605260 Review.
-
CRISPR-Cas inhibits plasmid transfer and immunizes bacteria against antibiotic resistance acquisition in manure.Appl Environ Microbiol. 2024 Sep 18;90(9):e0087624. doi: 10.1128/aem.00876-24. Epub 2024 Aug 19. Appl Environ Microbiol. 2024. PMID: 39158272 Free PMC article.
Cited by
-
Carbapenem Resistance in Acinetobacter baumannii: Mechanisms, Therapeutics, and Innovations.Microorganisms. 2025 Jun 27;13(7):1501. doi: 10.3390/microorganisms13071501. Microorganisms. 2025. PMID: 40732008 Free PMC article. Review.
-
Frontiers in superbug management: innovating approaches to combat antimicrobial resistance.Arch Microbiol. 2025 Feb 14;207(3):60. doi: 10.1007/s00203-025-04262-x. Arch Microbiol. 2025. PMID: 39953143 Review.
References
-
- Global Antimicrobial Resistance and Use Surveillance System (GLASS) Report: 2022. [(accessed on 13 October 2024)]. Available online: https://www.who.int/publications/i/item/9789240062702.
Publication types
LinkOut - more resources
Full Text Sources

