Novel Variant of New Delhi Metallo-Beta-Lactamase (blaNDM-60) Discovered in a Clinical Strain of Escherichia coli from the United Arab Emirates: An Emerging Challenge in Antimicrobial Resistance
- PMID: 39766548
- PMCID: PMC11672588
- DOI: 10.3390/antibiotics13121158
Novel Variant of New Delhi Metallo-Beta-Lactamase (blaNDM-60) Discovered in a Clinical Strain of Escherichia coli from the United Arab Emirates: An Emerging Challenge in Antimicrobial Resistance
Abstract
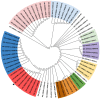
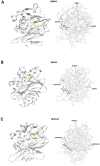
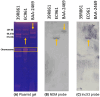
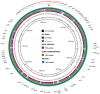
Background/Objectives: Carbapenem resistance poses a significant health threat. This study reports the first detection and characterization of a novel variant of New Delhi metallo-β-lactamase (blaNDM-60) in Escherichia coli from the United Arab Emirates (UAE), including its genetic context and relationship to global strains. Methods: NDM-60-producing E. coli was isolated from a rectal swab during routine screening. Characterization involved whole-genome sequencing, antimicrobial susceptibility testing, and comparative genomic analysis with 66 known NDM variants. Core genome analysis was performed against 42 global E. coli strains, including the single other reported NDM-60-positive isolate. Results: The strain demonstrated extensive drug resistance, including resistance to novel β-lactam/β-lactamase inhibitor combinations, notably taniborbactam. NDM-60 differs from the closely related NDM-5 by a single amino acid substitution (Asp202Asn) and two amino acid substitutions (Val88Leu and Met154Leu) compared to NDM-1. NDM-60 is located on a nonconjugative IncX3 plasmid. The strain belongs to sequence type 940 (ST940). Phylogenetic analysis revealed high diversity among the global ST940 strains, which carry a plethora of resistance genes and originated from humans, animals, and the environment from diverse geographic locations. Conclusions: NDM-60 emergence in the UAE represents a significant evolution in carbapenemase diversity. Its presence on a nonconjugative plasmid may limit spread; however, its extensive resistance profile is concerning. Further studies are needed to determine the prevalence, dissemination, and clinical impact of NDM-60. NDM evolution underscores the ongoing challenge in managing antimicrobial resistance and the critical importance of vigilant molecular surveillance. It also highlights the pressing demand to discover new antibiotics to fight resistant bacteria.
Keywords: Escherichia coli; NDM-60; New Delhi metallo-β-lactamase; United Arab Emirates; carbapenem resistance; whole-genome sequencing.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- Ma J., Song X., Li M., Yu Z., Cheng W., Yu Z., Zhang W., Zhang Y., Shen A., Sun H., et al. Global Spread of Carbapenem-Resistant Enterobacteriaceae: Epidemiological Features, Resistance Mechanisms, Detection and Therapy. Microbiol. Res. 2023;266:127249. doi: 10.1016/j.micres.2022.127249. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

