Embleporicin: A Novel Class I Lanthipeptide from the Actinobacteria Embleya sp. NF3
- PMID: 39766569
- PMCID: PMC11672506
- DOI: 10.3390/antibiotics13121179
Embleporicin: A Novel Class I Lanthipeptide from the Actinobacteria Embleya sp. NF3
Abstract
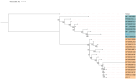
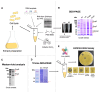
Genome mining has emerged as a revolutionary tool for discovering new ribosomally synthesized and post-translationally modified peptides (RiPPs) in various genomes. Recently, these approaches have been used to detect and explore unique environments as sources of RiPP-producing microorganisms, particularly focusing on endophytic microorganisms found in medicinal plants. Some endophytic actinobacteria, especially strains of Streptomyces, are notable examples of peptide producers, as specific biosynthetic clusters encode them. To uncover the genetic potential of these organisms, we analyzed the genome of the endophytic actinobacterium Embleya sp. NF3 using genome mining and bioinformatics tools. Our analysis led to the identification of a putative class I lanthipeptide. We cloned the core biosynthetic genes of this putative lanthipeptide, named embleporicin, and expressed them in vitro using a cell-free protein system (CFPS). The resulting product demonstrated antimicrobial activity against Micrococcus luteus ATCC 9341. This represents the first RiPP reported in the genus Embleya and the first actinobacterial lanthipeptide produced through cell-free technology.
Keywords: actinobacterium Embleya sp. NF3; antimicrobials; cell-free expression; genome mining; lanthipeptides.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- Arnison P.G., Bibb M.J., Bierbaum G., Bowers A.A., Bugni T.S., Bulaj G., Camarero J.A., Campopiano D.J., Challis G.L., Clardy J., et al. Ribosomally Synthesized and Post-Translationally Modified Peptide Natural Products: Overview and Recommendations for a Universal Nomenclature. Nat. Prod. Rep. 2013;30:108. doi: 10.1039/C2NP20085F. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

